Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance
- PMID: 34099649
- PMCID: PMC8184782
- DOI: 10.1038/s41467-021-23474-5
Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance
Abstract
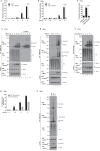
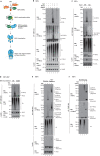
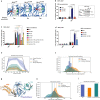
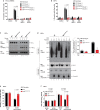
Necroptosis is a lytic, inflammatory form of cell death that not only contributes to pathogen clearance but can also lead to disease pathogenesis. Necroptosis is triggered by RIPK3-mediated phosphorylation of MLKL, which is thought to initiate MLKL oligomerisation, membrane translocation and membrane rupture, although the precise mechanism is incompletely understood. Here, we show that K63-linked ubiquitin chains are attached to MLKL during necroptosis and that ubiquitylation of MLKL at K219 significantly contributes to the cytotoxic potential of phosphorylated MLKL. The K219R MLKL mutation protects animals from necroptosis-induced skin damage and renders cells resistant to pathogen-induced necroptosis. Mechanistically, we show that ubiquitylation of MLKL at K219 is required for higher-order assembly of MLKL at membranes, facilitating its rupture and necroptosis. We demonstrate that K219 ubiquitylation licenses MLKL activity to induce lytic cell death, suggesting that necroptotic clearance of pathogens as well as MLKL-dependent pathologies are influenced by the ubiquitin-signalling system.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

