Reprogramming of microRNA expression via E2F1 downregulation promotes Salmonella infection both in infected and bystander cells
- PMID: 34099666
- PMCID: PMC8184997
- DOI: 10.1038/s41467-021-23593-z
Reprogramming of microRNA expression via E2F1 downregulation promotes Salmonella infection both in infected and bystander cells
Abstract
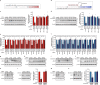
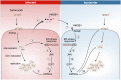
Cells infected with pathogens can contribute to clearing infections by releasing signals that instruct neighbouring cells to mount a pro-inflammatory cytokine response, or by other mechanisms that reduce bystander cells' susceptibility to infection. Here, we show the opposite effect: epithelial cells infected with Salmonella Typhimurium secrete host factors that facilitate the infection of bystander cells. We find that the endoplasmic reticulum stress response is activated in both infected and bystander cells, and this leads to activation of JNK pathway, downregulation of transcription factor E2F1, and consequent reprogramming of microRNA expression in a time-dependent manner. These changes are not elicited by infection with other bacterial pathogens, such as Shigella flexneri or Listeria monocytogenes. Remarkably, the protein HMGB1 present in the secretome of Salmonella-infected cells is responsible for the activation of the IRE1 branch of the endoplasmic reticulum stress response in non-infected, neighbouring cells. Furthermore, E2F1 downregulation and the associated microRNA alterations promote Salmonella replication within infected cells and prime bystander cells for more efficient infection.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

