Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection
- PMID: 34099924
- PMCID: PMC8245201
- DOI: 10.1038/s41591-021-01371-0
Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection
Abstract
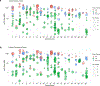
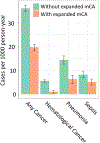
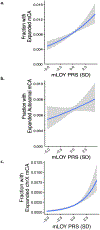
Age is the dominant risk factor for infectious diseases, but the mechanisms linking age to infectious disease risk are incompletely understood. Age-related mosaic chromosomal alterations (mCAs) detected from genotyping of blood-derived DNA, are structural somatic variants indicative of clonal hematopoiesis, and are associated with aberrant leukocyte cell counts, hematological malignancy, and mortality. Here, we show that mCAs predispose to diverse types of infections. We analyzed mCAs from 768,762 individuals without hematological cancer at the time of DNA acquisition across five biobanks. Expanded autosomal mCAs were associated with diverse incident infections (hazard ratio (HR) 1.25; 95% confidence interval (CI) = 1.15-1.36; P = 1.8 × 10-7), including sepsis (HR 2.68; 95% CI = 2.25-3.19; P = 3.1 × 10-28), pneumonia (HR 1.76; 95% CI = 1.53-2.03; P = 2.3 × 10-15), digestive system infections (HR 1.51; 95% CI = 1.32-1.73; P = 2.2 × 10-9) and genitourinary infections (HR 1.25; 95% CI = 1.11-1.41; P = 3.7 × 10-4). A genome-wide association study of expanded mCAs identified 63 loci, which were enriched at transcriptional regulatory sites for immune cells. These results suggest that mCAs are a marker of impaired immunity and confer increased predisposition to infections.
Figures













Update of
-
Hematopoietic mosaic chromosomal alterations and risk for infection among 767,891 individuals without blood cancer.Res Sq [Preprint]. 2020 Nov 16:rs.3.rs-100817. doi: 10.21203/rs.3.rs-100817/v1. Res Sq. 2020. Update in: Nat Med. 2021 Jun;27(6):1012-1024. doi: 10.1038/s41591-021-01371-0. PMID: 33236004 Free PMC article. Updated. Preprint.
Comment in
-
The vicious and virtuous circles of clonal hematopoiesis.Nat Med. 2021 Jun;27(6):949-950. doi: 10.1038/s41591-021-01396-5. Nat Med. 2021. PMID: 34099925 No abstract available.
References
-
- Gardner ID The effect of aging on susceptibility to infection. Rev Infect Dis 2, 801–810 (1980). - PubMed
-
- Gavazzi G & Krause KH Ageing and infection. Lancet Infect Dis 2, 659–666 (2002). - PubMed
-
- Franceschi C, Bonafe M & Valensin S Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine 18, 1717–1720 (2000). - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- F30 HL149180/HL/NHLBI NIH HHS/United States
- DP5 OD029586/OD/NIH HHS/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- R01 HL148565/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL128914/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- R01 HG006855/HG/NHGRI NIH HHS/United States
- DP2 ES030554/ES/NIEHS NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
- R01 MH104964/MH/NIMH NIH HHS/United States
- R01 HL151283/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- T32 GM136651/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

