Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study
- PMID: 34100002
- PMCID: PMC8172149
- DOI: 10.1016/S2666-5247(21)00090-2
Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study
Abstract
Background: Microbiological characterisation of co-infections and secondary infections in patients with COVID-19 is lacking, and antimicrobial use is high. We aimed to describe microbiologically confirmed co-infections and secondary infections, and antimicrobial use, in patients admitted to hospital with COVID-19.
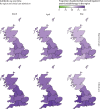
Methods: The International Severe Acute Respiratory and Emerging Infections Consortium (ISARIC) WHO Clinical Characterisation Protocol UK (CCP-UK) study is an ongoing, prospective cohort study recruiting inpatients from 260 hospitals in England, Scotland, and Wales, conducted by the ISARIC Coronavirus Clinical Characterisation Consortium. Patients with a confirmed or clinician-defined high likelihood of SARS-CoV-2 infection were eligible for inclusion in the ISARIC WHO CCP-UK study. For this specific study, we excluded patients with a recorded negative SARS-CoV-2 test result and those without a recorded outcome at 28 days after admission. Demographic, clinical, laboratory, therapeutic, and outcome data were collected using a prespecified case report form. Organisms considered clinically insignificant were excluded.
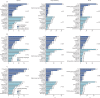
Findings: We analysed data from 48 902 patients admitted to hospital between Feb 6 and June 8, 2020. The median patient age was 74 years (IQR 59-84) and 20 786 (42·6%) of 48 765 patients were female. Microbiological investigations were recorded for 8649 (17·7%) of 48 902 patients, with clinically significant COVID-19-related respiratory or bloodstream culture results recorded for 1107 patients. 762 (70·6%) of 1080 infections were secondary, occurring more than 2 days after hospital admission. Staphylococcus aureus and Haemophilus influenzae were the most common pathogens causing respiratory co-infections (diagnosed ≤2 days after admission), with Enterobacteriaceae and S aureus most common in secondary respiratory infections. Bloodstream infections were most frequently caused by Escherichia coli and S aureus. Among patients with available data, 13 390 (37·0%) of 36 145 had received antimicrobials in the community for this illness episode before hospital admission and 39 258 (85·2%) of 46 061 patients with inpatient antimicrobial data received one or more antimicrobials at some point during their admission (highest for patients in critical care). We identified frequent use of broad-spectrum agents and use of carbapenems rather than carbapenem-sparing alternatives.
Interpretation: In patients admitted to hospital with COVID-19, microbiologically confirmed bacterial infections are rare, and more likely to be secondary infections. Gram-negative organisms and S aureus are the predominant pathogens. The frequency and nature of antimicrobial use are concerning, but tractable targets for stewardship interventions exist.
Funding: National Institute for Health Research (NIHR), UK Medical Research Council, Wellcome Trust, UK Department for International Development, Bill & Melinda Gates Foundation, EU Platform for European Preparedness Against (Re-)emerging Epidemics, NIHR Health Protection Research Unit (HPRU) in Emerging and Zoonotic Infections at University of Liverpool, and NIHR HPRU in Respiratory Infections at Imperial College London.
© 2021 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license.
Conflict of interest statement
All authors declare support from the NIHR, the Medical Research Council (MRC), the NIHR Health Protection Research Unit (HPRU) in Emerging and Zoonotic Infections at University of Liverpool, the NIHR HPRU in Respiratory Infections at Imperial College London, the NIHR Biomedical Research Centre (BRC) at Imperial College London, and the NIHR Clinical Research Network, for the submitted work. ABD reports grants from the UK Department of Health and Social Care (DHSC), during the conduct of the study, and grants from Wellcome Trust, outside the submitted work. PJMO reports personal fees from consultancies (GlaxoSmithKline, Janssen, Bavarian Nordic, Pfizer, and Cepheid) and from the European Respiratory Society, grants from MRC, MRC Global Challenge Research Fund, the EU, NIHR BRC, MRC–GlaxoSmithKline, Wellcome Trust, NIHR (HPRU in Respiratory Infection), and is an NIHR senior investigator outside the submitted work. PJMO's role as President of the British Society for Immunology was unpaid but travel and accommodation at some meetings was provided by the Society. JKB reports grants from MRC. MGS reports grants from DHSC, NIHR UK, MRC, HPRU in Emerging and Zoonotic Infections, and University of Liverpool, during the conduct of the study, and is chair of the scientific advisory board and a minority share holder at Integrum Scientific, outside the submitted work.
Figures



Comment in
-
COVID-19: stewardship of diagnostic tests for bacterial co-infection.Lancet Microbe. 2021 Nov;2(11):e570. doi: 10.1016/S2666-5247(21)00238-X. Epub 2021 Aug 26. Lancet Microbe. 2021. PMID: 34467256 Free PMC article. No abstract available.
References
-
- WHO . World Health Organization; Geneva: 2021. Clinical management of COVID-19: living guidance. Jan 25, 2021.
-
- National Institutes of Health Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. 2020. https://www.covid19treatmentguidelines.nih.gov/ - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12014/8/MRC_/Medical Research Council/United Kingdom
- MR/S032304/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/12/MRC_/Medical Research Council/United Kingdom
- G0701652/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/10/MRC_/Medical Research Council/United Kingdom
- MC_PC_19026/MRC_/Medical Research Council/United Kingdom
- 205228/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_19059/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/9/MRC_/Medical Research Council/United Kingdom
- MC_PC_19025/MRC_/Medical Research Council/United Kingdom
- MC_PC_15001/MRC_/Medical Research Council/United Kingdom
- 215091/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

