Synthesis and biological evaluation of 4-phenoxy-phenyl isoxazoles as novel acetyl-CoA carboxylase inhibitors
- PMID: 34100310
- PMCID: PMC8205039
- DOI: 10.1080/14756366.2021.1936514
Synthesis and biological evaluation of 4-phenoxy-phenyl isoxazoles as novel acetyl-CoA carboxylase inhibitors
Abstract
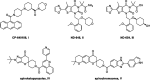
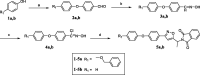
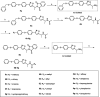
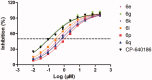
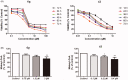
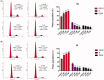
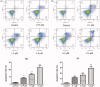
Acetyl-CoA carboxylase (ACC) is a crucial enzyme in fatty acid metabolism, which plays a major role in the occurrence and development of certain tumours. Herein, one potential ACC inhibitor (6a) was identified through high-throughput virtual screening (HTVS), and a series of 4-phenoxy-phenyl isoxazoles were synthesised for structure-activity relationship (SAR) studies. Among these compounds, 6g exhibited the most potent ACC inhibitory activity (IC50=99.8 nM), which was comparable to that of CP-640186. Moreover, the antiproliferation assay revealed that compound 6l exhibited the strongest cytotoxicity, with IC50 values of 0.22 µM (A549), 0.26 µM (HepG2), and 0.21 µM (MDA-MB-231), respectively. The preliminary mechanistic studies on 6g and 6l suggested that the compounds decreased the malonyl-CoA levels, arrested the cell cycle at the G0/G1 phase, and induced apoptosis in MDA-MB-231 cells. Overall, these results indicated that the 4-phenoxy-phenyl isoxazoles are potential for further study in cancer therapeutics as ACC inhibitors.
Keywords: Acetyl-CoA carboxylase; antitumor; apoptosis; cell cycle; docking.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
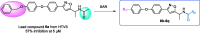
Similar articles
-
Synthesis and in vitro evaluation of novel spiroketopyrazoles as acetyl-CoA carboxylase inhibitors and potential antitumor agents.Eur J Med Chem. 2021 Feb 15;212:113036. doi: 10.1016/j.ejmech.2020.113036. Epub 2020 Nov 27. Eur J Med Chem. 2021. PMID: 33276990
-
Synthesis, Biological Evaluation and Molecular Docking Studies of Piperidinylpiperidines and Spirochromanones Possessing Quinoline Moieties as Acetyl-CoA Carboxylase Inhibitors.Molecules. 2015 Sep 7;20(9):16221-34. doi: 10.3390/molecules200916221. Molecules. 2015. PMID: 26370948 Free PMC article.
-
Isozyme-nonselective N-substituted bipiperidylcarboxamide acetyl-CoA carboxylase inhibitors reduce tissue malonyl-CoA concentrations, inhibit fatty acid synthesis, and increase fatty acid oxidation in cultured cells and in experimental animals.J Biol Chem. 2003 Sep 26;278(39):37099-111. doi: 10.1074/jbc.M304481200. Epub 2003 Jul 3. J Biol Chem. 2003. PMID: 12842871
-
Recent development in acetyl-CoA carboxylase inhibitors and their potential as novel drugs.Future Med Chem. 2020 Mar;12(6):533-561. doi: 10.4155/fmc-2019-0312. Epub 2020 Feb 12. Future Med Chem. 2020. PMID: 32048880 Review.
-
Acetyl-CoA carboxylase (ACC) as a therapeutic target for metabolic syndrome and recent developments in ACC1/2 inhibitors.Expert Opin Investig Drugs. 2019 Oct;28(10):917-930. doi: 10.1080/13543784.2019.1657825. Epub 2019 Aug 29. Expert Opin Investig Drugs. 2019. PMID: 31430206 Review.
Cited by
-
Metabolomics-driven approaches for identifying therapeutic targets in drug discovery.MedComm (2020). 2024 Nov 11;5(11):e792. doi: 10.1002/mco2.792. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39534557 Free PMC article. Review.
References
-
- Braig S. Chemical genetics in tumor lipogenesis. Biotechnol Adv 2018;36:1724–9. - PubMed
-
- Rohrig F, Schulze A.. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer 2016;16:732–49. - PubMed
-
- Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
-
- Ngoi NYL, Eu JQ, Hirpara J, et al. . Targeting cell metabolism as cancer therapy. Antioxid Redox Signal 2020;32:285–308. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
