Computational Model To Quantify the Growth of Antibiotic-Resistant Bacteria in Wastewater
- PMID: 34100640
- PMCID: PMC8579810
- DOI: 10.1128/mSystems.00360-21
Computational Model To Quantify the Growth of Antibiotic-Resistant Bacteria in Wastewater
Abstract
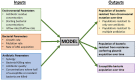
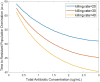
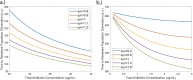
Although wastewater and sewage systems are known to be significant reservoirs of antibiotic-resistant bacterial populations and periodic outbreaks of drug-resistant infection, there is little quantitative understanding of the drivers behind resistant population growth in these settings. In order to fill this gap in quantitative understanding of the development of antibiotic-resistant infections in wastewater, we have developed a mathematical model synthesizing many known drivers of antibiotic resistance in these settings to help predict the growth of resistant populations in different environmental scenarios. A number of these drivers of drug-resistant infection outbreak, including antibiotic residue concentration, antibiotic interaction, chromosomal mutation, and horizontal gene transfer, have not previously been integrated into a single computational model. We validated the outputs of the model with quantitative studies conducted on the eVOLVER continuous culture platform. Our integrated model shows that low levels of antibiotic residues present in wastewater can lead to increased development of resistant populations and that the dominant mechanism of resistance acquisition in these populations is horizontal gene transfer rather than acquisition of chromosomal mutations. Additionally, we found that synergistic antibiotics at low concentrations lead to increased resistant population growth. These findings, consistent with recent experimental and field studies, provide new quantitative knowledge on the evolution of antibiotic-resistant bacterial reservoirs, and the model developed herein can be adapted for use as a prediction tool in public health policy making, particularly in low-income settings where water sanitation issues remain widespread and disease outbreaks continue to undermine public health efforts. IMPORTANCE The rate at which antimicrobial resistance (AMR) has developed and spread throughout the world has increased in recent years, and according to the Review on Antimicrobial Resistance in 2014, it is suggested that the current rate will lead to AMR-related deaths of several million people by 2050 (Review on Antimicrobial Resistance, Tackling a Crisis for the Health and Wealth of Nations, 2014). One major reservoir of resistant bacterial populations that has been linked to outbreaks of drug-resistant bacterial infections but is not well understood is in wastewater settings, where antibiotic pollution is often present. Using ordinary differential equations incorporating several known drivers of resistance in wastewater, we find that interactions between antibiotic residues and horizontal gene transfer significantly affect the growth of resistant bacterial reservoirs.
Keywords: antibiotic resistance; mathematical modeling; wastewater.
Figures







References
-
- Nadimpalli ML, Marks SJ, Montealegre MC, Gilman RH, Pajuelo MJ, Saito M, Tsukayama P, Njenga SM, Kiiru J, Swarthout J, Islam MA, Julian TR, Pickering AJ. 2020. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat Microbiol 5:787–795. doi:10.1038/s41564-020-0722-0. - DOI - PubMed
-
- Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, Shaheen G, Qureshi S, Yousafzai MT, Saleem MK, Hasan Z, Dougan G, Hasan R. 2018. Emergence of an extensively drug-resistant Salmonella enterica serovar typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9:e00105-18. doi:10.1128/mBio.00105-18. - DOI - PMC - PubMed
-
- Gothwal R, Shashidhar T. 2015. Antibiotic pollution in the environment: a review. CLEAN Soil Air Water 43:479–489. doi:10.1002/clen.201300989. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

