FOXA1 overexpression suppresses interferon signaling and immune response in cancer
- PMID: 34101624
- PMCID: PMC8279591
- DOI: 10.1172/JCI147025
FOXA1 overexpression suppresses interferon signaling and immune response in cancer
Abstract
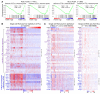
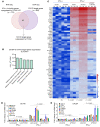
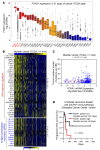
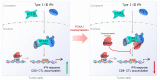
Androgen receptor-positive prostate cancer (PCa) and estrogen receptor-positive luminal breast cancer (BCa) are generally less responsive to immunotherapy compared with certain tumor types such as melanoma. However, the underlying mechanisms are not fully elucidated. In this study, we found that FOXA1 overexpression inversely correlated with interferon (IFN) signature and antigen presentation gene expression in PCa and BCa patients. FOXA1 bound the STAT2 DNA-binding domain and suppressed STAT2 DNA-binding activity, IFN signaling gene expression, and cancer immune response independently of the transactivation activity of FOXA1 and its mutations detected in PCa and BCa. Increased FOXA1 expression promoted cancer immuno- and chemotherapy resistance in mice and PCa and BCa patients. These findings were also validated in bladder cancer expressing high levels of FOXA1. FOXA1 overexpression could be a prognostic factor to predict therapy resistance and a viable target to sensitize luminal PCa, BCa, and bladder cancer to immuno- and chemotherapy.
Keywords: Cancer; Cancer immunotherapy; Cell Biology; Molecular biology; Therapeutics.
Conflict of interest statement
Figures





References
-
- Wei SC, et al. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

