De novo and bi-allelic variants in AP1G1 cause neurodevelopmental disorder with developmental delay, intellectual disability, and epilepsy
- PMID: 34102099
- PMCID: PMC8322935
- DOI: 10.1016/j.ajhg.2021.05.007
De novo and bi-allelic variants in AP1G1 cause neurodevelopmental disorder with developmental delay, intellectual disability, and epilepsy
Abstract
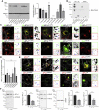
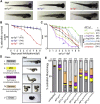
Adaptor protein (AP) complexes mediate selective intracellular vesicular trafficking and polarized localization of somatodendritic proteins in neurons. Disease-causing alleles of various subunits of AP complexes have been implicated in several heritable human disorders, including intellectual disabilities (IDs). Here, we report two bi-allelic (c.737C>A [p.Pro246His] and c.1105A>G [p.Met369Val]) and eight de novo heterozygous variants (c.44G>A [p.Arg15Gln], c.103C>T [p.Arg35Trp], c.104G>A [p.Arg35Gln], c.229delC [p.Gln77Lys∗11], c.399_400del [p.Glu133Aspfs∗37], c.747G>T [p.Gln249His], c.928-2A>C [p.?], and c.2459C>G [p.Pro820Arg]) in AP1G1, encoding gamma-1 subunit of adaptor-related protein complex 1 (AP1γ1), associated with a neurodevelopmental disorder (NDD) characterized by mild to severe ID, epilepsy, and developmental delay in eleven families from different ethnicities. The AP1γ1-mediated adaptor complex is essential for the formation of clathrin-coated intracellular vesicles. In silico analysis and 3D protein modeling simulation predicted alteration of AP1γ1 protein folding for missense variants, which was consistent with the observed altered AP1γ1 levels in heterologous cells. Functional studies of the recessively inherited missense variants revealed no apparent impact on the interaction of AP1γ1 with other subunits of the AP-1 complex but rather showed to affect the endosome recycling pathway. Knocking out ap1g1 in zebrafish leads to severe morphological defect and lethality, which was significantly rescued by injection of wild-type AP1G1 mRNA and not by transcripts encoding the missense variants. Furthermore, microinjection of mRNAs with de novo missense variants in wild-type zebrafish resulted in severe developmental abnormalities and increased lethality. We conclude that de novo and bi-allelic variants in AP1G1 are associated with neurodevelopmental disorder in diverse populations.
Keywords: AP-1 complex; AP1G1; Pakistani families; developmental delay; epilepsy; exome sequencing; genetic heterogeneity; intellectual disabilities; neurodevelopment disorder.
Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hirst J., Robinson M.S. Clathrin and adaptors. Biochim. Biophys. Acta. 1998;1404:173–193. - PubMed
-
- Robinson M.S. Forty Years of Clathrin-coated Vesicles. Traffic. 2015;16:1210–1238. - PubMed
-
- Ohno H. Clathrin-associated adaptor protein complexes. J. Cell Sci. 2006;119:3719–3721. - PubMed
-
- Traub L.M., Kornfeld S., Ungewickell E. Different Domains of AP1 Adaptor complex are required for Golgi membrane binding and clathrine recruitment. J. Biol. Chem. 1995;270:4933–4942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

