Resolving the graft ischemia-reperfusion injury during liver transplantation at the single cell resolution
- PMID: 34103479
- PMCID: PMC8187624
- DOI: 10.1038/s41419-021-03878-3
Resolving the graft ischemia-reperfusion injury during liver transplantation at the single cell resolution
Abstract
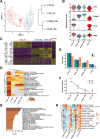
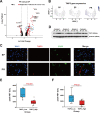
Ischemia-reperfusion injury (IRI) remains the major reason for impaired donor graft function and increased mortality post-liver transplantation. The mechanism of IRI involves multiple pathophysiological processes and numerous types of cells. However, a systematic and comprehensive single-cell transcriptional profile of intrahepatic cells during liver transplantation is still unclear. We performed a single-cell transcriptome analysis of 14,313 cells from liver tissues collected from pre-procurement, at the end of preservation and 2 h post-reperfusion. We made detailed annotations of mononuclear phagocyte, endothelial cell, NK/T, B and plasma cell clusters, and we described the dynamic changes of the transcriptome of these clusters during IRI and the interaction between mononuclear phagocyte clusters and other cell clusters. In addition, we found that TNFAIP3 interacting protein 3 (TNIP3), specifically and highly expressed in Kupffer cell clusters post-reperfusion, may have a protective effect on IRI. In summary, our study provides the first dynamic transcriptome map of intrahepatic cell clusters during liver transplantation at single-cell resolution.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Discovery of Endothelial-Monocyte Crosstalk in Ischemic-Reperfusion Injury Following Liver Transplantation Based on Integration of Single-Cell RNA and Transcriptome RNA Sequencing.J Cell Mol Med. 2025 Feb;29(4):e70336. doi: 10.1111/jcmm.70336. J Cell Mol Med. 2025. PMID: 39993960 Free PMC article.
-
Genome-wide DNA methylation and transcriptomic analysis of liver tissues subjected to early ischemia/reperfusion injury upon human liver transplantation.Ann Hepatol. 2024 Jul-Aug;29(4):101506. doi: 10.1016/j.aohep.2024.101506. Epub 2024 May 6. Ann Hepatol. 2024. PMID: 38710471
-
Ischaemia reperfusion injury in liver transplantation: Cellular and molecular mechanisms.Liver Int. 2019 May;39(5):788-801. doi: 10.1111/liv.14091. Epub 2019 Apr 2. Liver Int. 2019. PMID: 30843314 Free PMC article. Review.
-
Genetic effect of ischemia-reperfusion injury upon primary graft dysfunction and chronic lung allograft dysfunction in lung transplantation: evidence based on transcriptome data.Transpl Immunol. 2022 Apr;71:101556. doi: 10.1016/j.trim.2022.101556. Epub 2022 Feb 22. Transpl Immunol. 2022. PMID: 35202801
-
Role of hepatic stellate cells in liver ischemia-reperfusion injury.Front Immunol. 2022 Jul 28;13:891868. doi: 10.3389/fimmu.2022.891868. eCollection 2022. Front Immunol. 2022. PMID: 35967364 Free PMC article. Review.
Cited by
-
The Impact of Biliary Injury on the Recurrence of Biliary Cancer and Benign Disease after Liver Transplantation: Risk Factors and Mechanisms.Cancers (Basel). 2024 Aug 7;16(16):2789. doi: 10.3390/cancers16162789. Cancers (Basel). 2024. PMID: 39199562 Free PMC article. Review.
-
Inhibition of cysteine-serine-rich nuclear protein 1 ameliorates ischemia-reperfusion injury during liver transplantation in an MAPK-dependent manner.Mol Biomed. 2024 Jun 21;5(1):22. doi: 10.1186/s43556-024-00185-z. Mol Biomed. 2024. PMID: 38902590 Free PMC article.
-
Advanced omics approaches in liver transplant settings: current applications and future prospectives.Front Immunol. 2025 Jul 11;16:1564248. doi: 10.3389/fimmu.2025.1564248. eCollection 2025. Front Immunol. 2025. PMID: 40718477 Free PMC article. Review.
-
Sterile Pancreas Inflammation during Preservation and after Transplantation.Int J Mol Sci. 2023 Feb 27;24(5):4636. doi: 10.3390/ijms24054636. Int J Mol Sci. 2023. PMID: 36902067 Free PMC article. Review.
-
Dynamic immune recovery process after liver transplantation revealed by single-cell multi-omics analysis.Innovation (Camb). 2024 Mar 1;5(3):100599. doi: 10.1016/j.xinn.2024.100599. eCollection 2024 May 6. Innovation (Camb). 2024. PMID: 38510071 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

