Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers
- PMID: 34103486
- PMCID: PMC8187732
- DOI: 10.1038/s41467-021-23507-z
Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers
Abstract
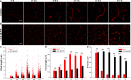
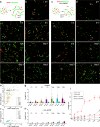
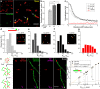
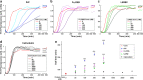
Several cell-surface receptors for neurotoxic forms of amyloid-β (Aβ) have been described, but their molecular interactions with Aβ assemblies and their relative contributions to mediating Alzheimer's disease pathology have remained uncertain. Here, we used super-resolution microscopy to directly visualize Aβ-receptor interactions at the nanometer scale. We report that one documented Aβ receptor, PrPC, specifically inhibits the polymerization of Aβ fibrils by binding to the rapidly growing end of each fibril, thereby blocking polarized elongation at that end. PrPC binds neurotoxic oligomers and protofibrils in a similar fashion, suggesting that it may recognize a common, end-specific, structural motif on all of these assemblies. Finally, two other Aβ receptors, FcγRIIb and LilrB2, affect Aβ fibril growth in a manner similar to PrPC. Our results suggest that receptors may trap Aβ oligomers and protofibrils on the neuronal surface by binding to a common molecular determinant on these assemblies, thereby initiating a neurotoxic signal.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Systematic and standardized comparison of reported amyloid-β receptors for sufficiency, affinity, and Alzheimer's disease relevance.J Biol Chem. 2019 Apr 12;294(15):6042-6053. doi: 10.1074/jbc.RA118.006252. Epub 2019 Feb 20. J Biol Chem. 2019. PMID: 30787106 Free PMC article.
-
Cellular prion protein targets amyloid-β fibril ends via its C-terminal domain to prevent elongation.J Biol Chem. 2017 Oct 13;292(41):16858-16871. doi: 10.1074/jbc.M117.789990. Epub 2017 Aug 23. J Biol Chem. 2017. PMID: 28842494 Free PMC article.
-
Protofibril-Fibril Interactions Inhibit Amyloid Fibril Assembly by Obstructing Secondary Nucleation.Angew Chem Int Ed Engl. 2021 Feb 8;60(6):3016-3021. doi: 10.1002/anie.202010098. Epub 2020 Dec 11. Angew Chem Int Ed Engl. 2021. PMID: 33095508 Free PMC article.
-
The Binding Receptors of Aβ: an Alternative Therapeutic Target for Alzheimer's Disease.Mol Neurobiol. 2016 Jan;53(1):455-471. doi: 10.1007/s12035-014-8994-0. Epub 2014 Dec 4. Mol Neurobiol. 2016. PMID: 25465238 Review.
-
Protofibrils of Amyloid-β are Important Targets of a Disease-Modifying Approach for Alzheimer's Disease.Int J Mol Sci. 2020 Jan 31;21(3):952. doi: 10.3390/ijms21030952. Int J Mol Sci. 2020. PMID: 32023927 Free PMC article. Review.
Cited by
-
Alzheimer's disease: a mini-review for the clinician.Front Neurol. 2023 Jun 22;14:1178588. doi: 10.3389/fneur.2023.1178588. eCollection 2023. Front Neurol. 2023. PMID: 37426432 Free PMC article. Review.
-
Unveiling the Potential of Polyphenols as Anti-Amyloid Molecules in Alzheimer's Disease.Curr Neuropharmacol. 2023;21(4):787-807. doi: 10.2174/1570159X20666221010113812. Curr Neuropharmacol. 2023. PMID: 36221865 Free PMC article. Review.
-
Single-molecule fluorescence imaging and deep learning reveal highly heterogeneous aggregation of amyloid-β 42.Proc Natl Acad Sci U S A. 2022 Mar 22;119(12):e2116736119. doi: 10.1073/pnas.2116736119. Epub 2022 Mar 15. Proc Natl Acad Sci U S A. 2022. PMID: 35290118 Free PMC article.
-
Structural details of amyloid β oligomers in complex with human prion protein as revealed by solid-state MAS NMR spectroscopy.J Biol Chem. 2021 Jan-Jun;296:100499. doi: 10.1016/j.jbc.2021.100499. Epub 2021 Mar 3. J Biol Chem. 2021. PMID: 33667547 Free PMC article.
-
MicroRNAs as Potential Orchestrators of Alzheimer's Disease-Related Pathologies: Insights on Current Status and Future Possibilities.Front Aging Neurosci. 2021 Oct 12;13:743573. doi: 10.3389/fnagi.2021.743573. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34712129 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

