Molecular evidence of SARS-CoV-2 in New York before the first pandemic wave
- PMID: 34103497
- PMCID: PMC8187428
- DOI: 10.1038/s41467-021-23688-7
Molecular evidence of SARS-CoV-2 in New York before the first pandemic wave
Abstract
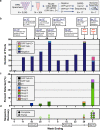
Numerous reports document the spread of SARS-CoV-2, but there is limited information on its introduction before the identification of a local case. This may lead to incorrect assumptions when modeling viral origins and transmission. Here, we utilize a sample pooling strategy to screen for previously undetected SARS-CoV-2 in de-identified, respiratory pathogen-negative nasopharyngeal specimens from 3,040 patients across the Mount Sinai Health System in New York. The patients had been previously evaluated for respiratory symptoms or influenza-like illness during the first 10 weeks of 2020. We identify SARS-CoV-2 RNA from specimens collected as early as 25 January 2020, and complete SARS-CoV-2 genome sequences from multiple pools of samples collected between late February and early March, documenting an increase prior to the later surge. Our results provide evidence of sporadic SARS-CoV-2 infections a full month before both the first officially documented case and emergence of New York as a COVID-19 epicenter in March 2020.
Conflict of interest statement
R.S. is VP of Technology Development and a stockholder at Sema4, a Mount Sinai Venture. This work, however, was conducted solely at Icahn School of Medicine at Mount Sinai. Otherwise, the authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

