Identifying and characterizing high-risk clusters in a heterogeneous ICU population with deep embedded clustering
- PMID: 34103544
- PMCID: PMC8187398
- DOI: 10.1038/s41598-021-91297-x
Identifying and characterizing high-risk clusters in a heterogeneous ICU population with deep embedded clustering
Abstract
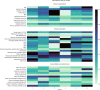
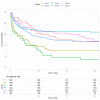
Critically ill patients constitute a highly heterogeneous population, with seemingly distinct patients having similar outcomes, and patients with the same admission diagnosis having opposite clinical trajectories. We aimed to develop a machine learning methodology that identifies and provides better characterization of patient clusters at high risk of mortality and kidney injury. We analysed prospectively collected data including co-morbidities, clinical examination, and laboratory parameters from a minimally-selected population of 743 patients admitted to the ICU of a Dutch hospital between 2015 and 2017. We compared four clustering methodologies and trained a classifier to predict and validate cluster membership. The contribution of different variables to the predicted cluster membership was assessed using SHapley Additive exPlanations values. We found that deep embedded clustering yielded better results compared to the traditional clustering algorithms. The best cluster configuration was achieved for 6 clusters. All clusters were clinically recognizable, and differed in in-ICU, 30-day, and 90-day mortality, as well as incidence of acute kidney injury. We identified two high mortality risk clusters with at least 60%, 40%, and 30% increased. ICU, 30-day and 90-day mortality, and a low risk cluster with 25-56% lower mortality risk. This machine learning methodology combining deep embedded clustering and variable importance analysis, which we made publicly available, is a possible solution to challenges previously encountered by clustering analyses in heterogeneous patient populations and may help improve the characterization of risk groups in critical care.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

