A patient-based medaka alg2 mutant as a model for hypo-N-glycosylation
- PMID: 34106226
- PMCID: PMC8217707
- DOI: 10.1242/dev.199385
A patient-based medaka alg2 mutant as a model for hypo-N-glycosylation
Abstract
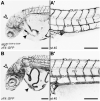
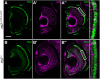
Defects in the evolutionarily conserved protein-glycosylation machinery during embryonic development are often fatal. Consequently, congenital disorders of glycosylation (CDG) in human are rare. We modelled a putative hypomorphic mutation described in an alpha-1,3/1,6-mannosyltransferase (ALG2) index patient (ALG2-CDG) to address the developmental consequences in the teleost medaka (Oryzias latipes). We observed specific, multisystemic, late-onset phenotypes, closely resembling the patient's syndrome, prominently in the facial skeleton and in neuronal tissue. Molecularly, we detected reduced levels of N-glycans in medaka and in the patient's fibroblasts. This hypo-N-glycosylation prominently affected protein abundance. Proteins of the basic glycosylation and glycoprotein-processing machinery were over-represented in a compensatory response, highlighting the regulatory topology of the network. Proteins of the retinal phototransduction machinery, conversely, were massively under-represented in the alg2 model. These deficiencies relate to a specific failure to maintain rod photoreceptors, resulting in retinitis pigmentosa characterized by the progressive loss of these photoreceptors. Our work has explored only the tip of the iceberg of N-glycosylation-sensitive proteins, the function of which specifically impacts on cells, tissues and organs. Taking advantage of the well-described human mutation has allowed the complex interplay of N-glycosylated proteins and their contribution to development and disease to be addressed.
Keywords: CDG; Disease model; Glycosylation; Human hypomorphic mutations; Medaka; Retinitis pigmentosa.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures






References
-
- Ba-Abbad, R., Leys, M., Wang, X., Chakarova, C., Waseem, N., Carss, K. J., Raymond, F. L., Bujakowska, K. M., Pierce, E. A., Mahroo, O. A.et al. (2018). Clinical features of a retinopathy associated with a dominant allele of the RGR gene. Invest. Ophthalmol. Vis. Sci. 59, 4812-4820. 10.1167/iovs.18-25061 - DOI - PMC - PubMed
-
- Cantagrel, V., Lefeber, D. J., Ng, B. G., Guan, Z., Silhavy, J. L., Bielas, S. L., Lehle, L., Hombauer, H., Adamowicz, M., Swiezewska, E.et al. (2010). SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 142, 203-217. 10.1016/j.cell.2010.06.001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

