Serum Analyte Profiles Associated With Crohn's Disease and Disease Location
- PMID: 34106269
- PMCID: PMC8730700
- DOI: 10.1093/ibd/izab123
Serum Analyte Profiles Associated With Crohn's Disease and Disease Location
Abstract
Background: Crohn's disease (CD) can affect any segment of the digestive tract but is most often localized in the ileal, ileocolonic, and colorectal regions of the intestines. It is believed that the chronic inflammation in CD is a result of an imbalance between the epithelial barrier, the immune system, and the intestinal microbiota. The aim of the study was to identify circulating markers associated with CD and/or disease location in CD patients.
Methods: We tested 49 cytokines, chemokines, and growth factors in serum samples from 300 patients with CD and 300 controls. After quality control, analyte levels were tested for association with CD and disease location.
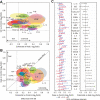
Results: We identified 13 analytes that were higher in CD patients relative to healthy controls and that remained significant after conservative Bonferroni correction (P < 0.0015). In particular, CXCL9, CXCL1, and interleukin IL-6 had the greatest effect and were highly significant (P < 5 × 10-7). We also identified 9 analytes that were associated with disease location, with VEGF, IL-12p70, and IL-6 being elevated in patients with colorectal disease (P < 3 × 10-4).
Conclusions: Multiple serum analytes are elevated in CD. These implicate the involvement of multiple cell types from the immune, epithelial, and endothelial systems, suggesting that circulating analytes reflect the inflammatory processes that are ongoing within the gut. Moreover, the identification of distinct profiles according to disease location supports the existence of a biological difference between ileal and colonic CD, consistent with previous genetic and clinical observations.
Keywords: Crohn’s disease; disease location; serum biomarkers.
© 2021 Crohn’s & Colitis Foundation. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380:1590–1605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

