Epigenetic Plasticity Enables CNS-Trafficking of EBV-infected B Lymphocytes
- PMID: 34106998
- PMCID: PMC8216538
- DOI: 10.1371/journal.ppat.1009618
Epigenetic Plasticity Enables CNS-Trafficking of EBV-infected B Lymphocytes
Abstract
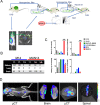
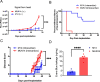
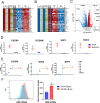
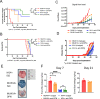
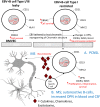
Subpopulations of B-lymphocytes traffic to different sites and organs to provide diverse and tissue-specific functions. Here, we provide evidence that epigenetic differences confer a neuroinvasive phenotype. An EBV+ B cell lymphoma cell line (M14) with low frequency trafficking to the CNS was neuroadapted to generate a highly neuroinvasive B-cell population (MUN14). MUN14 B cells efficiently infiltrated the CNS within one week and produced neurological pathologies. We compared the gene expression profiles of viral and cellular genes using RNA-Seq and identified one viral (EBNA1) and several cellular gene candidates, including secreted phosphoprotein 1/osteopontin (SPP1/OPN), neuron navigator 3 (NAV3), CXCR4, and germinal center-associated signaling and motility protein (GCSAM) that were selectively upregulated in MUN14. ATAC-Seq and ChIP-qPCR revealed that these gene expression changes correlated with epigenetic changes at gene regulatory elements. The neuroinvasive phenotype could be attenuated with a neutralizing antibody to OPN, confirming the functional role of this protein in trafficking EBV+ B cells to the CNS. These studies indicate that B-cell trafficking to the CNS can be acquired by epigenetic adaptations and provide a new model to study B-cell neuroinvasion associated CNS lymphoma and autoimmune disease of the CNS, including multiple sclerosis (MS).
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Paul M. Lieberman declares his role as a founder of Vironika, LLC as a Conflict of Interest that is managed by the Wistar Institute. No other authors have any conflicts to disclose.
Figures


References
-
- Young LS, Arrand JR, Murray PG. EBV gene expression and regulation. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, et al.., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge 2007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

