Replicative senescence dictates the emergence of disease-associated microglia and contributes to Aβ pathology
- PMID: 34107254
- PMCID: PMC8206957
- DOI: 10.1016/j.celrep.2021.109228
Replicative senescence dictates the emergence of disease-associated microglia and contributes to Aβ pathology
Abstract
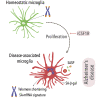
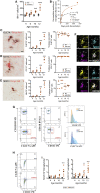
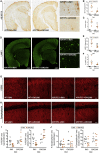
The sustained proliferation of microglia is a key hallmark of Alzheimer's disease (AD), accelerating its progression. Here, we aim to understand the long-term impact of the early and prolonged microglial proliferation observed in AD, hypothesizing that extensive and repeated cycling would engender a distinct transcriptional and phenotypic trajectory. We show that the early and sustained microglial proliferation seen in an AD-like model promotes replicative senescence, characterized by increased βgal activity, a senescence-associated transcriptional signature, and telomere shortening, correlating with the appearance of disease-associated microglia (DAM) and senescent microglial profiles in human post-mortem AD cases. The prevention of early microglial proliferation hinders the development of senescence and DAM, impairing the accumulation of Aβ, as well as associated neuritic and synaptic damage. Overall, our results indicate that excessive microglial proliferation leads to the generation of senescent DAM, which contributes to early Aβ pathology in AD.
Keywords: APP/PS1; Alzheimer's disease; CSF1R; disease-associated microglia (DAM).
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Alliot F., Godin I., Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain Res. 1999;117:145–152. - PubMed
-
- Askew K., Gomez-Nicola D. A story of birth and death: insights into the formation and dynamics of the microglial population. Brain Behav. Immun. 2018;69:9–17. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

