Expanding the taxonomic range in the fecal metagenome
- PMID: 34107881
- PMCID: PMC8188691
- DOI: 10.1186/s12859-021-04212-6
Expanding the taxonomic range in the fecal metagenome
Abstract
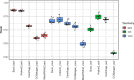
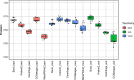
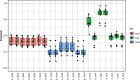
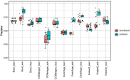
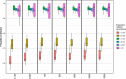
Background: Except for bacteria, the taxonomic diversity of the human fecal metagenome has not been widely studied, despite the potential importance of viruses and eukaryotes. Widely used bioinformatic tools contain limited numbers of non-bacterial species in their databases compared to available genomic sequences and their methodologies do not favour classification of rare sequences which may represent only a small fraction of their parent genome. In seeking to optimise identification of non-bacterial species, we evaluated five widely-used metagenome classifier programs (BURST, Kraken2, Centrifuge, MetaPhlAn2 and CCMetagen) for their ability to correctly assign and count simulations of bacterial, viral and eukaryotic DNA sequence reads, including the effect of taxonomic order of analysis of bacteria, viruses and eukaryotes and the effect of sequencing depth.
Results: We found that the precision of metagenome classifiers varied significantly between programs and between taxonomic groups. When classifying viruses and eukaryotes, ordering the analysis such that bacteria were classified first significantly improved classification precision. Increasing sequencing depth decreased classification precision and did not improve recall of rare species.
Conclusions: Choice of metagenome classifier program can have a marked effect on results with respect to precision of species assignment in different taxonomic groups. The order of taxonomic classification can markedly improve precision. Increasing sequencing depth can decrease classification precision and yields diminishing returns in probability of species detection.
Keywords: Benchmarking; Classifier; Eukaryotes; Metagenomics; Viruses.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

