GNINA 1.0: molecular docking with deep learning
- PMID: 34108002
- PMCID: PMC8191141
- DOI: 10.1186/s13321-021-00522-2
GNINA 1.0: molecular docking with deep learning
Abstract
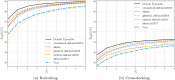
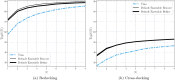
Molecular docking computationally predicts the conformation of a small molecule when binding to a receptor. Scoring functions are a vital piece of any molecular docking pipeline as they determine the fitness of sampled poses. Here we describe and evaluate the 1.0 release of the Gnina docking software, which utilizes an ensemble of convolutional neural networks (CNNs) as a scoring function. We also explore an array of parameter values for Gnina 1.0 to optimize docking performance and computational cost. Docking performance, as evaluated by the percentage of targets where the top pose is better than 2Å root mean square deviation (Top1), is compared to AutoDock Vina scoring when utilizing explicitly defined binding pockets or whole protein docking. GNINA, utilizing a CNN scoring function to rescore the output poses, outperforms AutoDock Vina scoring on redocking and cross-docking tasks when the binding pocket is defined (Top1 increases from 58% to 73% and from 27% to 37%, respectively) and when the whole protein defines the binding pocket (Top1 increases from 31% to 38% and from 12% to 16%, respectively). The derived ensemble of CNNs generalizes to unseen proteins and ligands and produces scores that correlate well with the root mean square deviation to the known binding pose. We provide the 1.0 version of GNINA under an open source license for use as a molecular docking tool at https://github.com/gnina/gnina .
Keywords: Deep learning; Molecular docking; Structure-based drug design.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures











References
-
- Muegge I. A knowledge-based scoring function for protein-ligand interactions: probing the reference state. Perspect Drug Discov Design. 2000;20(1):99–114. doi: 10.1023/A:1008729005958. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

