Applications of single-cell sequencing in cancer research: progress and perspectives
- PMID: 34108022
- PMCID: PMC8190846
- DOI: 10.1186/s13045-021-01105-2
Applications of single-cell sequencing in cancer research: progress and perspectives
Abstract
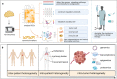
Single-cell sequencing, including genomics, transcriptomics, epigenomics, proteomics and metabolomics sequencing, is a powerful tool to decipher the cellular and molecular landscape at a single-cell resolution, unlike bulk sequencing, which provides averaged data. The use of single-cell sequencing in cancer research has revolutionized our understanding of the biological characteristics and dynamics within cancer lesions. In this review, we summarize emerging single-cell sequencing technologies and recent cancer research progress obtained by single-cell sequencing, including information related to the landscapes of malignant cells and immune cells, tumor heterogeneity, circulating tumor cells and the underlying mechanisms of tumor biological behaviors. Overall, the prospects of single-cell sequencing in facilitating diagnosis, targeted therapy and prognostic prediction among a spectrum of tumors are bright. In the near future, advances in single-cell sequencing will undoubtedly improve our understanding of the biological characteristics of tumors and highlight potential precise therapeutic targets for patients.
Keywords: Circulating tumor cells; Heterogeneity; Microenvironment; Single-cell sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

