Multiomics analysis reveals CT83 is the most specific gene for triple negative breast cancer and its hypomethylation is oncogenic in breast cancer
- PMID: 34108519
- PMCID: PMC8190062
- DOI: 10.1038/s41598-021-91290-4
Multiomics analysis reveals CT83 is the most specific gene for triple negative breast cancer and its hypomethylation is oncogenic in breast cancer
Abstract
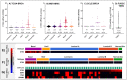
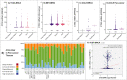
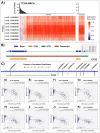
Triple-negative breast cancer (TNBC) is a highly aggressive breast cancer (BrC) subtype lacking effective therapeutic targets currently. The development of multi-omics databases facilities the identification of core genes for TNBC. Using TCGA-BRCA and METABRIC datasets, we identified CT83 as the most TNBC-specific gene. By further integrating FUSCC-TNBC, CCLE, TCGA pan-cancer, Expression Atlas, and Human Protein Atlas datasets, we found CT83 is frequently activated in TNBC and many other cancers, while it is always silenced in non-TNBC, 120 types of normal non-testis tissues, and 18 types of blood cells. Notably, according to the TCGA-BRCA methylation data, hypomethylation on chromosome X 116,463,019 to 116,463,039 is significantly correlated with the abnormal activation of CT83 in BrC. Using Kaplan-Meier Plotter, we demonstrated that activated CT83 is significantly associated with unfavorably overall survival in BrC and worse outcomes in some other cancers. Furthermore, GSEA suggested that the abnormal activation of CT83 in BrC is probably oncogenic by triggering the activation of cell cycle signaling. Meanwhile, we also noticed copy number variations and mutations of CT83 are quite rare in any cancer type, and its role in immune infiltration is not significant. In summary, we highlighted the significance of CT83 for TNBC and presented a comprehensive bioinformatics strategy for single-gene analysis in cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

