Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability
- PMID: 34108662
- PMCID: PMC7611218
- DOI: 10.1038/s41556-021-00692-z
Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability
Erratum in
-
Publisher Correction: Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability.Nat Cell Biol. 2021 Jul;23(7):809. doi: 10.1038/s41556-021-00720-y. Nat Cell Biol. 2021. PMID: 34163038 Free PMC article. No abstract available.
Abstract
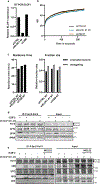
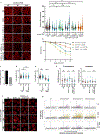
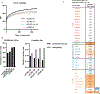
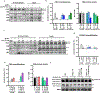
Correct transcription is crucial for life. However, DNA damage severely impedes elongating RNA polymerase II, causing transcription inhibition and transcription-replication conflicts. Cells are equipped with intricate mechanisms to counteract the severe consequence of these transcription-blocking lesions. However, the exact mechanism and factors involved remain largely unknown. Here, using a genome-wide CRISPR-Cas9 screen, we identified the elongation factor ELOF1 as an important factor in the transcription stress response following DNA damage. We show that ELOF1 has an evolutionarily conserved role in transcription-coupled nucleotide excision repair (TC-NER), where it promotes recruitment of the TC-NER factors UVSSA and TFIIH to efficiently repair transcription-blocking lesions and resume transcription. Additionally, ELOF1 modulates transcription to protect cells against transcription-mediated replication stress, thereby preserving genome stability. Thus, ELOF1 protects the transcription machinery from DNA damage via two distinct mechanisms.
Conflict of interest statement
Competing interests:
Authors declare no competing interests.
Figures











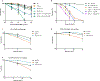
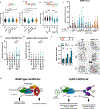
Comment in
-
The ELOF(1)ant in the room of TCR.Nat Cell Biol. 2021 Jun;23(6):584-586. doi: 10.1038/s41556-021-00698-7. Nat Cell Biol. 2021. PMID: 34108661 No abstract available.
References
-
- Lans H, Hoeijmakers JHJ, Vermeulen W & Marteijn JA The DNA damage response to transcription stress. Nat Rev Mol Cell Biol 20, 766–784 (2019). - PubMed
-
- Gaillard H & Aguilera A Transcription as a Threat to Genome Integrity. Annu Rev Biochem 85, 291–317 (2016). - PubMed
-
- Laugel V Cockayne syndrome: the expanding clinical and mutational spectrum. Mech Ageing Dev 134, 161–170 (2013). - PubMed
Online methods References:
-
- Yesbolatova A, Natsume T, Hayashi K. i. & Kanemaki MT Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods 164–165, 73–80 (2019). - PubMed
-
- Natsume T, Kiyomitsu T, Saga Y & Kanemaki MT Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep 15, 210–218 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

