ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation
- PMID: 34108663
- PMCID: PMC8890769
- DOI: 10.1038/s41556-021-00688-9
ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation
Abstract
Cells employ transcription-coupled repair (TCR) to eliminate transcription-blocking DNA lesions. DNA damage-induced binding of the TCR-specific repair factor CSB to RNA polymerase II (RNAPII) triggers RNAPII ubiquitylation of a single lysine (K1268) by the CRL4CSA ubiquitin ligase. How CRL4CSA is specifically directed towards K1268 is unknown. Here, we identify ELOF1 as the missing link that facilitates RNAPII ubiquitylation, a key signal for the assembly of downstream repair factors. This function requires its constitutive interaction with RNAPII close to K1268, revealing ELOF1 as a specificity factor that binds and positions CRL4CSA for optimal RNAPII ubiquitylation. Drug-genetic interaction screening also revealed a CSB-independent pathway in which ELOF1 prevents R-loops in active genes and protects cells against DNA replication stress. Our study offers key insights into the molecular mechanisms of TCR and provides a genetic framework of the interplay between transcriptional stress responses and DNA replication.
Conflict of interest statement
Figures




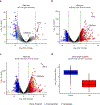

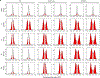
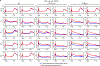

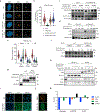


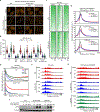


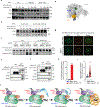


Comment in
-
The ELOF(1)ant in the room of TCR.Nat Cell Biol. 2021 Jun;23(6):584-586. doi: 10.1038/s41556-021-00698-7. Nat Cell Biol. 2021. PMID: 34108661 No abstract available.
References
-
- Brueckner F, Hennecke U, Carell T & Cramer P CPD damage recognition by transcribing RNA polymerase II. Science 315, 859–862 (2007). - PubMed
-
- Nakazawa Y et al. Ubiquitination of DNA Damage-Stalled RNAPII Promotes Transcription-Coupled Repair. Cell 180, 1228–1244.e1224 (2020). - PubMed
-
- Nakazawa Y et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet 44, 586–592 (2012). - PubMed
-
- Schwertman P et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44, 598–602 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

