Origins and Evolution of the Primate Hepatitis B Virus
- PMID: 34108947
- PMCID: PMC8180572
- DOI: 10.3389/fmicb.2021.653684
Origins and Evolution of the Primate Hepatitis B Virus
Abstract
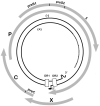
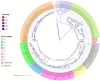
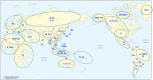
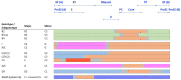
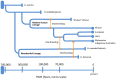
Recent interest in the origins and subsequent evolution of the hepatitis B virus (HBV) has strengthened with the discovery of ancient HBV sequences in fossilized remains of humans dating back to the Neolithic period around 7,000 years ago. Metagenomic analysis identified a number of African non-human primate HBV sequences in the oldest samples collected, indicating that human HBV may have at some stage, evolved in Africa following zoonotic transmissions from higher primates. Ancestral genotype A and D isolates were also discovered from the Bronze Age, not in Africa but rather Eurasia, implying a more complex evolutionary and migratory history for HBV than previously recognized. Most full-length ancient HBV sequences exhibited features of inter genotypic recombination, confirming the importance of recombination and the mutation rate of the error-prone viral replicase as drivers for successful HBV evolution. A model for the origin and evolution of HBV is proposed, which includes multiple cross-species transmissions and favors subsequent recombination events that result in a pathogen and can successfully transmit and cause persistent infection in the primate host.
Keywords: ancient DNA; evolution; genotype; hepatitis B virus; human migration.
Copyright © 2021 Locarnini, Littlejohn and Yuen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Beard K. C. (2004). The Hunt for the Dawn Monkey: Unearthing the Origins of Monkeys, Apes and Humans. LA: University of California Press.
Publication types
LinkOut - more resources
Full Text Sources

