Lineage-specific mechanisms and drivers of breast cancer chemoresistance revealed by 3D biomimetic culture
- PMID: 34109737
- PMCID: PMC8847989
- DOI: 10.1002/1878-0261.13037
Lineage-specific mechanisms and drivers of breast cancer chemoresistance revealed by 3D biomimetic culture
Abstract
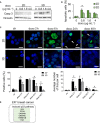
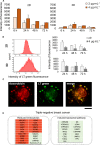
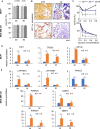
To improve the success rate of current preclinical drug trials, there is a growing need for more complex and relevant models that can help predict clinical resistance to anticancer agents. Here, we present a three-dimensional (3D) technology, based on biomimetic collagen scaffolds, that enables the modeling of the tumor hypoxic state and the prediction of in vivo chemotherapy responses in terms of efficacy, molecular alterations, and emergence of resistance mechanisms. The human breast cancer cell lines MDA-MB-231 (triple negative) and MCF-7 (luminal A) were treated with scaling doses of doxorubicin in monolayer cultures, 3D collagen scaffolds, or orthotopically transplanted murine models. Lineage-specific resistance mechanisms were revealed by the 3D tumor model. Reduced drug uptake, increased drug efflux, and drug lysosomal confinement were observed in triple-negative MDA-MB-231 cells. In luminal A MCF-7 cells, the selection of a drug-resistant subline from parental cells with deregulation of p53 pathways occurred. These cells were demonstrated to be insensitive to DNA damage. Transcriptome analysis was carried out to identify differentially expressed genes (DEGs) in treated cells. DEG evaluation in breast cancer patients demonstrated their potential role as predictive biomarkers. High expression of the transporter associated with antigen processing 1 (TAP1) and the tumor protein p53-inducible protein 3 (TP53I3) was associated with shorter relapse in patients affected by ER+ breast tumor. Likewise, the same clinical outcome was associated with high expression of the lysosomal-associated membrane protein 1 LAMP1 in triple-negative breast cancer. Hypoxia inhibition by resveratrol treatment was found to partially re-sensitize cells to doxorubicin treatment. Our model might improve preclinical in vitro analysis for the translation of anticancer compounds as it provides: (a) more accurate data on drug efficacy and (b) enhanced understanding of resistance mechanisms and molecular drivers.
Keywords: 3D models; DNA repair; breast cancer; doxorubicin; drug resistance; lysosomes.
© 2021 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Yan X, Zhou L, Wu Z, Wang X, Chen X, Yang F, Guo Y, Wu M, Chen Y, Li W et al. (2019) High throughput scaffold‐based 3D micro‐tumor array for efficient drug screening and chemosensitivity testing. Biomaterials 198, 167–179. - PubMed
-
- Hay M, Thomas DW, Craighead JL, Economides C & Rosenthal J (2014) Clinical development success rates for investigational drugs. Nat Biotechnol 32, 40–51. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

