SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors
- PMID: 34110264
- PMCID: PMC8316110
- DOI: 10.1128/JVI.00266-21
SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors
Abstract
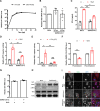
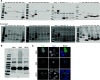
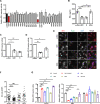
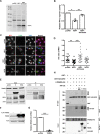
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic. While previous studies have shown that several SARS-CoV-2 proteins can antagonize the interferon (IFN) response, some of the mechanisms by which they do so are not well understood. In this study, we describe two novel mechanisms by which SARS-CoV-2 blocks the IFN pathway. Type I IFNs and IFN-stimulated genes (ISGs) were poorly induced during SARS-CoV-2 infection, and once infection was established, cells were highly resistant to ectopic induction of IFNs and ISGs. Levels of two key IFN signaling pathway components, Tyk2 and STAT2, were significantly lower in SARS-CoV-2-infected cells. Expression of nonstructural protein 1 (NSP1) or nucleocapsid in the absence of other viral proteins was sufficient to block IFN induction, but only NSP1 was able to inhibit IFN signaling. Mapping studies suggest that NSP1 prevents IFN induction in part by blocking IRF3 phosphorylation. In addition, NSP1-induced depletion of Tyk2 and STAT2 dampened ISG induction. Together, our data provide new insights into how SARS-CoV-2 successfully evades the IFN system to establish infection. IMPORTANCE SARS-CoV-2 is the causative agent of COVID-19, a serious disease that can have a myriad of symptoms from loss of taste and smell to pneumonia and hypercoagulation. The rapid spread of SARS-CoV-2 can be attributed in part to asymptomatic transmission, where infected individuals shed large amounts of virus before the onset of disease. This is likely due to the ability of SARS-CoV-2 to effectively suppress the innate immune system, including the IFN response. Indeed, we show that the IFN response is efficiently blocked during SARS-CoV-2 infection, a process that is mediated in large part by nonstructural protein 1 and nucleocapsid. Our study provides new insights on how SARS-CoV-2 evades the IFN response to successfully establish infection. These findings should be considered for the development and administration of therapeutics against SARS-CoV-2.
Keywords: NSP1; SARS-CoV-2; innate immunity; type I interferons.
Figures
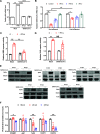
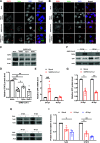
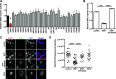
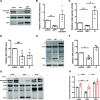
Similar articles
-
Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options.Viruses. 2022 Jun 8;14(6):1247. doi: 10.3390/v14061247. Viruses. 2022. PMID: 35746718 Free PMC article. Review.
-
Evasion of Type I Interferon by SARS-CoV-2.Cell Rep. 2020 Oct 6;33(1):108234. doi: 10.1016/j.celrep.2020.108234. Epub 2020 Sep 19. Cell Rep. 2020. PMID: 32979938 Free PMC article.
-
SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response.Cells. 2021 Mar 2;10(3):530. doi: 10.3390/cells10030530. Cells. 2021. PMID: 33801464 Free PMC article.
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Antagonism of Type I Interferon by Severe Acute Respiratory Syndrome Coronavirus 2.J Interferon Cytokine Res. 2020 Dec;40(12):543-548. doi: 10.1089/jir.2020.0214. J Interferon Cytokine Res. 2020. PMID: 33337934 Free PMC article. Review.
Cited by
-
Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options.Viruses. 2022 Jun 8;14(6):1247. doi: 10.3390/v14061247. Viruses. 2022. PMID: 35746718 Free PMC article. Review.
-
The Wnt/β-catenin pathway is important for replication of SARS-CoV-2 and other pathogenic RNA viruses.Npj Viruses. 2024 Feb 21;2(1):6. doi: 10.1038/s44298-024-00018-4. Npj Viruses. 2024. PMID: 40295745 Free PMC article.
-
Distinct airway epithelial immune responses after infection with SARS-CoV-2 compared to H1N1.Mucosal Immunol. 2022 May;15(5):952-963. doi: 10.1038/s41385-022-00545-4. Epub 2022 Jul 15. Mucosal Immunol. 2022. PMID: 35840680 Free PMC article.
-
Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2.Nat Commun. 2021 Dec 7;12(1):7092. doi: 10.1038/s41467-021-27318-0. Nat Commun. 2021. PMID: 34876592 Free PMC article.
-
Antiviral responses versus virus-induced cellular shutoff: a game of thrones between influenza A virus NS1 and SARS-CoV-2 Nsp1.Front Cell Infect Microbiol. 2024 Feb 5;14:1357866. doi: 10.3389/fcimb.2024.1357866. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38375361 Free PMC article. Review.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA, Public Health–Seattle and King County and CDC COVID-19 Investigation Team . 2020. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med 382:2081–2090. 10.1056/NEJMoa2008457. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Canada Excellence Research Chair/Gouvernement du Canada | Canadian Institutes of Health Research (CIHR)
- Rapid Response/U of A | Li Ka Shing Institute of Virology, University of Alberta (LKSIV)
- OV3- 172302/Gouvernement du Canada | Canadian Institutes of Health Research (CIHR)
- PJT 159442/Gouvernement du Canada | Canadian Institutes of Health Research (CIHR)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

