Humanization of antibodies using a machine learning approach on large-scale repertoire data
- PMID: 34110413
- PMCID: PMC8760955
- DOI: 10.1093/bioinformatics/btab434
Humanization of antibodies using a machine learning approach on large-scale repertoire data
Abstract
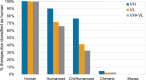
Motivation: Monoclonal antibody (mAb) therapeutics are often produced from non-human sources (typically murine), and can therefore generate immunogenic responses in humans. Humanization procedures aim to produce antibody therapeutics that do not elicit an immune response and are safe for human use, without impacting efficacy. Humanization is normally carried out in a largely trial-and-error experimental process. We have built machine learning classifiers that can discriminate between human and non-human antibody variable domain sequences using the large amount of repertoire data now available.
Results: Our classifiers consistently outperform the current best-in-class model for distinguishing human from murine sequences, and our output scores exhibit a negative relationship with the experimental immunogenicity of existing antibody therapeutics. We used our classifiers to develop a novel, computational humanization tool, Hu-mAb, that suggests mutations to an input sequence to reduce its immunogenicity. For a set of therapeutic antibodies with known precursor sequences, the mutations suggested by Hu-mAb show substantial overlap with those deduced experimentally. Hu-mAb is therefore an effective replacement for trial-and-error humanization experiments, producing similar results in a fraction of the time.
Availability and implementation: Hu-mAb (humanness scoring and humanization) is freely available to use at opig.stats.ox.ac.uk/webapps/humab.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures



References
-
- Chirino A.J. et al. (2004) Minimizingthe immunogenicity of protein therapeutics. Drug Discov. Today, 9, 82–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

