Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis
- PMID: 34111196
- PMCID: PMC8191978
- DOI: 10.1371/journal.pone.0253007
Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis
Abstract
Background: Diagnosis of COVID-19 in symptomatic patients and screening of populations for SARS-CoV-2 infection require access to straightforward, low-cost and high-throughput testing. The recommended nasopharyngeal swab tests are limited by the need of trained professionals and specific consumables and this procedure is poorly accepted as a screening method In contrast, saliva sampling can be self-administered.
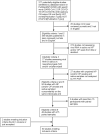
Methods: In order to compare saliva and nasopharyngeal/oropharyngeal samples for the detection of SARS-CoV-2, we designed a meta-analysis searching in PubMed up to December 29th, 2020 with the key words "(SARS-CoV-2 OR COVID-19 OR COVID19) AND (salivary OR saliva OR oral fluid)) NOT (review[Publication Type]) NOT (PrePrint[Publication Type])" applying the following criteria: records published in peer reviewed scientific journals, in English, with at least 15 nasopharyngeal/orapharyngeal swabs and saliva paired samples tested by RT-PCR, studies with available raw data including numbers of positive and negative tests with the two sampling methods. For all studies, concordance and sensitivity were calculated and then pooled in a random-effects model.
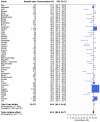
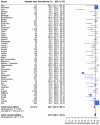
Findings: A total of 377 studies were retrieved, of which 50 were eligible, reporting on 16,473 pairs of nasopharyngeal/oropharyngeal and saliva samples. Meta-analysis showed high concordance, 92.5% (95%CI: 89.5-94.7), across studies and pooled sensitivities of 86.5% (95%CI: 83.4-89.1) and 92.0% (95%CI: 89.1-94.2) from saliva and nasopharyngeal/oropharyngeal swabs respectively. Heterogeneity across studies was 72.0% for saliva and 85.0% for nasopharyngeal/oropharyngeal swabs.
Interpretation: Our meta-analysis strongly suggests that saliva could be used for frequent testing of COVID-19 patients and "en masse" screening of populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-nove... (accessed Jan. 3, 2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

