Long non-coding RNA KIKAT/LINC01061 as a novel epigenetic regulator that relocates KDM4A on chromatin and modulates viral reactivation
- PMID: 34111227
- PMCID: PMC8219169
- DOI: 10.1371/journal.ppat.1009670
Long non-coding RNA KIKAT/LINC01061 as a novel epigenetic regulator that relocates KDM4A on chromatin and modulates viral reactivation
Abstract
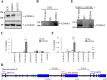
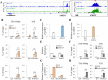
KDM4A is a histone lysine demethylase that has been described as an oncogene in various types of cancer. The importance of KDM4A-mediated epigenetic regulation in tumorigenesis is just emerging. Here, by using Kaposi's sarcoma associated herpesvirus (KSHV) as a screening model, we identified 6 oncogenic virus-induced long non-coding RNAs (lncRNAs) with the potential to open chromatin. RNA immunoprecipitation revealed KSHV-induced KDM4A-associated transcript (KIKAT)/LINC01061 as a binding partner of KDM4A. Integrated ChIP-seq and RNA-seq analysis showed that the KIKAT/LINC01061 interaction may mediate relocalization of KDM4A from the transcription start site (TSS) of the AMOT promoter region and transactivation of AMOT, an angiostatin binding protein that regulates endothelial cell migration. Knockdown of AMOT diminished the migration ability of uninfected SLK and iSLK-BAC16 cells in response to KIKAT/LINC01061 overexpression. Thus, we conclude that KIKAT/LINC01061 triggered shifting of KDM4A as a potential epigenetic mechanism regulating gene transactivation. Dysregulation of KIKAT/LINC01061 expression may represent a novel pathological mechanism contributing to KDM4A oncogenicity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures