KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin
- PMID: 34111513
- PMCID: PMC8324551
- DOI: 10.1016/j.canlet.2021.05.031
KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin
Abstract
Mutations in KRAS frequently occur in human cancer and are especially prevalent in pancreatic ductal adenocarcinoma (PDAC), where they have been shown to promote aggressive phenotypes. However, targeting this onco-protein has proven to be challenging, highlighting the need to further identify the various mechanisms used by KRAS to drive cancer progression. Here, we considered the role played by exosomes, a specific class of extracellular vesicles (EVs) derived from the endocytic cellular trafficking machinery, in mediating the ability of KRAS to promote cell survival. We found that exosomes isolated from the serum of PDAC patients, as well as from KRAS-transformed fibroblasts and pancreatic cancer cells, were all highly enriched in the cell survival protein Survivin. Exosomes containing Survivin, upon engaging serum-starved cells, strongly enhanced their survival. Moreover, they significantly compromised the effectiveness of the conventional chemotherapy drug paclitaxel, as well as a novel therapy that combines an ERK inhibitor with chloroquine, which is currently in clinical trials for PDAC. The survival benefits provided by oncogenic KRAS-derived exosomes were markedly reduced when depleted of Survivin using siRNA or upon treatment with the Survivin inhibitor YM155. Taken together, these findings demonstrate how KRAS mutations give rise to exosomes that provide a unique form of intercellular communication to promote cancer cell survival and therapy resistance, as well as raise interesting possibilities regarding their potential for serving as therapeutic targets and diagnostic markers for KRAS-dependent cancers.
Keywords: Exosome; Extracellular vesicle; KRAS; Pancreatic cancer; Survivin.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest
C.J.D. is a consultant/advisory board member for Anchiano Therapeutics, Deciphera Pharmaceuticals and Mirati Therapeutics. C.J.D. has received research funding support from SpringWorks Therapeutics, Mirati Therapeutics and Deciphera Pharmaceuticals, and has consulted for Eli Lilly, Jazz Therapeutics, Revolution Medicines, Ribometrix, Sanofi, and Turning Point Therapeutics.
Figures



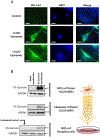


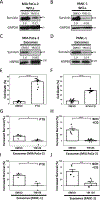

References
-
- Stephen AG, Esposito D, Bagni RG &McCormick F Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

