Lactobacillus rhamnosus GG Orchestrates an Antitumor Immune Response
- PMID: 34111601
- PMCID: PMC8463873
- DOI: 10.1016/j.jcmgh.2021.06.001
Lactobacillus rhamnosus GG Orchestrates an Antitumor Immune Response
Abstract
Background & aims: In colorectal cancer, approximately 95% of patients are refractory to immunotherapy because of low antitumor immune responses. Therefore, there is an exigent need to develop treatments that increase antitumor immune responses and decrease tumor burden to enhance immunotherapy.
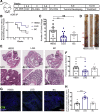
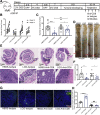
Methods: The gut microbiome has been described as a master modulator of immune responses. We administered the human commensal, Lactobacillus rhamnosus GG (LGG), to mice and characterized the changes in the gut immune landscape. Because the presence of lactobacilli in the gut microbiome has been linked with decreased tumor burden and antitumor immune responses, we also supplemented a genetic and a chemical model of murine intestinal cancer with LGG. For clinical relevance, we therapeutically administered LGG after tumors had formed. We also tested for the requirement of CD8 T cells in LGG-mediated modulation of gut tumor burden.
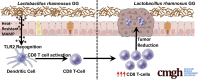
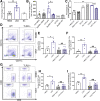
Results: We detected increased colonic CD8 T-cell responses specifically in LGG-supplemented mice. The CD8 T-cell induction was dependent on dendritic cell activation mediated via Toll-like receptor-2, thereby describing a novel mechanism in which a member of the human microbiome induces an intestinal CD8 T-cell response. We also show that LGG decreased tumor burden in the murine gut cancer models by a CD8 T-cell-dependent manner.
Conclusions: These data support the potential use of LGG to augment antitumor immune responses in colorectal cancer patients and ultimately for increasing the breadth and efficacy of immunotherapy.
Keywords: CD8 T Cells; CRC; Cancer; Dendritic Cells; LGG; Microbiome; Probiotics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. - PubMed
-
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012: global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. - PubMed
-
- Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. - PubMed
-
- Overman M.J., Lonardi S., Wong K.Y.M., Lenz H.-J., Gelsomino F., Aglietta M., Morse M.A., Van Cutsem E., McDermott R., Hill A., Sawyer M.B., Hendlisz A., Neyns B., Svrcek M., Moss R.A., Ledeine J.-M., Cao Z.A., Kamble S., Kopetz S., André T. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. - PubMed
-
- Overman M.J., Ernstoff M.S., Morse M.A. Where we stand with immunotherapy in colorectal cancer: deficient mismatch repair, proficient mismatch repair, and toxicity management. Am Soc Clin Oncol. 2018;38:239–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

