Rapid detection of identity-by-descent tracts for mega-scale datasets
- PMID: 34112768
- PMCID: PMC8192555
- DOI: 10.1038/s41467-021-22910-w
Rapid detection of identity-by-descent tracts for mega-scale datasets
Abstract
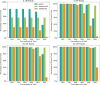
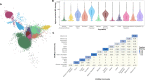
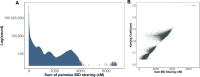
The ability to identify segments of genomes identical-by-descent (IBD) is a part of standard workflows in both statistical and population genetics. However, traditional methods for finding local IBD across all pairs of individuals scale poorly leading to a lack of adoption in very large-scale datasets. Here, we present iLASH, an algorithm based on similarity detection techniques that shows equal or improved accuracy in simulations compared to current leading methods and speeds up analysis by several orders of magnitude on genomic datasets, making IBD estimation tractable for millions of individuals. We apply iLASH to the PAGE dataset of ~52,000 multi-ethnic participants, including several founder populations with elevated IBD sharing, identifying IBD segments in ~3 minutes per chromosome compared to over 6 days for a state-of-the-art algorithm. iLASH enables efficient analysis of very large-scale datasets, as we demonstrate by computing IBD across the UK Biobank (~500,000 individuals), detecting 12.9 billion pairwise connections.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

