Enhanced mechanosensing of cells in synthetic 3D matrix with controlled biophysical dynamics
- PMID: 34112772
- PMCID: PMC8192531
- DOI: 10.1038/s41467-021-23120-0
Enhanced mechanosensing of cells in synthetic 3D matrix with controlled biophysical dynamics
Abstract
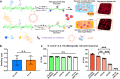
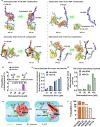
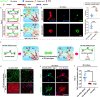
3D culture of cells in designer biomaterial matrices provides a biomimetic cellular microenvironment and can yield critical insights into cellular behaviours not available from conventional 2D cultures. Hydrogels with dynamic properties, achieved by incorporating either degradable structural components or reversible dynamic crosslinks, enable efficient cell adaptation of the matrix and support associated cellular functions. Herein we demonstrate that given similar equilibrium binding constants, hydrogels containing dynamic crosslinks with a large dissociation rate constant enable cell force-induced network reorganization, which results in rapid stellate spreading, assembly, mechanosensing, and differentiation of encapsulated stem cells when compared to similar hydrogels containing dynamic crosslinks with a low dissociation rate constant. Furthermore, the static and precise conjugation of cell adhesive ligands to the hydrogel subnetwork connected by such fast-dissociating crosslinks is also required for ultra-rapid stellate spreading (within 18 h post-encapsulation) and enhanced mechanosensing of stem cells in 3D. This work reveals the correlation between microscopic cell behaviours and the molecular level binding kinetics in hydrogel networks. Our findings provide valuable guidance to the design and evaluation of supramolecular biomaterials with cell-adaptable properties for studying cells in 3D cultures.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Briquez, P. S., Clegg, L. E., Martino, M. M., Mac Gabhann, F. & Hubbell, J. A. Design principles for therapeutic angiogenic materials. Nat. Rev. Mater.1, 15006 (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

