Characterization of metapopulation of Ellobium chinense through Pleistocene expansions and four covariate COI guanine-hotspots linked to G-quadruplex conformation
- PMID: 34112865
- PMCID: PMC8192772
- DOI: 10.1038/s41598-021-91675-5
Characterization of metapopulation of Ellobium chinense through Pleistocene expansions and four covariate COI guanine-hotspots linked to G-quadruplex conformation
Abstract
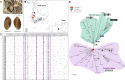
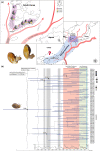
The land snail Ellobium chinense (L. Pfeiffer, 1855) (Eupulmonata, Ellobiida, Ellobiidae), which inhabits the salt marshes along the coastal areas of northwestern Pacific, is an endangered species on the IUCN Red List. Over recent decades, the population size of E. chinense has consistently decreased due to environmental interference caused by natural disasters and human activities. Here, we provide the first assessment of the genetic diversity and population genetic structures of northwestern Pacific E. chinense. The results analyzed with COI and microsatellites revealed that E. chinense population exhibit metapopulation characteristics, retaining under the influence of the Kuroshio warm currents through expansion of the Late-Middle and Late Pleistocene. We also found four phylogenetic groups, regardless of geographical distributions, which were easily distinguishable by four unidirectional and stepwise adenine-to-guanine transitions in COI (sites 207-282-354-420: A-A-A-A, A-A-G-A, G-A-G-A, and G-G-G-G). Additionally, the four COI hotspots were robustly connected with a high degree of covariance between them. We discuss the role of these covariate guanines which link to form four consecutive G-quadruplexes, and their possible beneficial effects under positive selection pressure.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Pfeiffer L. Descriptions of seven new species of land shells from the collection of H. cuming. Esq. Proc. Zool. Soc. Lond. 1864;32:603–605.
-
- Yoo JS. Korean Shells in Colour. IlJiSa Publishing Co.; 1976. pp. 1–196.
-
- Ministry of Oceans and Fisheries. Conservation and management of marine ecosystems act. Korea Ministry of Government Legislation. https://www.law.go.kr/LSW/eng/engLsSc.do?menuId=2&query=CONSERVATION+AND... (2017).
-
- Mable BK. Conservation of adaptive potential and functional diversity: Integrating old and new approaches. Conserv. Genet. 2019;20:89–100. doi: 10.1007/s10592-018-1129-9. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

