Macropinocytosis requires Gal-3 in a subset of patient-derived glioblastoma stem cells
- PMID: 34112916
- PMCID: PMC8192788
- DOI: 10.1038/s42003-021-02258-z
Macropinocytosis requires Gal-3 in a subset of patient-derived glioblastoma stem cells
Abstract
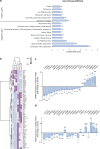
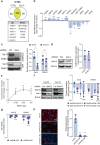
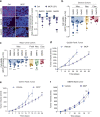
Recently, we involved the carbohydrate-binding protein Galectin-3 (Gal-3) as a druggable target for KRAS-mutant-addicted lung and pancreatic cancers. Here, using glioblastoma patient-derived stem cells (GSCs), we identify and characterize a subset of Gal-3high glioblastoma (GBM) tumors mainly within the mesenchymal subtype that are addicted to Gal-3-mediated macropinocytosis. Using both genetic and pharmacologic inhibition of Gal-3, we showed a significant decrease of GSC macropinocytosis activity, cell survival and invasion, in vitro and in vivo. Mechanistically, we demonstrate that Gal-3 binds to RAB10, a member of the RAS superfamily of small GTPases, and β1 integrin, which are both required for macropinocytosis activity and cell survival. Finally, by defining a Gal-3/macropinocytosis molecular signature, we could predict sensitivity to this dependency pathway and provide proof-of-principle for innovative therapeutic strategies to exploit this Achilles' heel for a significant and unique subset of GBM patients.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

