Function-Oriented Graphene Quantum Dots Probe for Single Cell in situ Sorting of Active Microorganisms in Environmental Samples
- PMID: 34113325
- PMCID: PMC8186282
- DOI: 10.3389/fmicb.2021.659111
Function-Oriented Graphene Quantum Dots Probe for Single Cell in situ Sorting of Active Microorganisms in Environmental Samples
Abstract
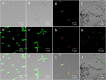
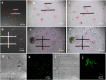
Functional microorganisms play a vital role in removing environmental pollutants because of their diverse metabolic capability. Herein, a function-oriented fluorescence resonance energy transfer (FRET)-based graphene quantum dots (GQDs-M) probe was developed for the specific identification and accurate sorting of azo-degrading functional bacteria in the original location of environmental samples for large-scale culturing. First, nitrogen-doped GQDs (GQDs-N) were synthesized using a bottom-up strategy. Then, a GQDs-M probe was synthesized based on bonding FRET-based GQDs-N to an azo dye, methyl red, and the quenched fluorescence was recovered upon cleavage of the azo bond. Bioimaging confirmed the specific recognition capability of GQDs-M upon incubation with the target bacteria or environmental samples. It is suggested that the estimation of environmental functional microbial populations based on bioimaging will be a new method for rapid preliminary assessment of environmental pollution levels. In combination with a visual single-cell sorter, the target bacteria in the environmental samples could be intuitively screened at the single-cell level in 17 bacterial strains, including the positive control Shewanella decolorationis S12, and were isolated from environmental samples. All of these showed an azo degradation function, indicating the high accuracy of the single-cell sorting strategy using the GQDs-M. Furthermore, among the bacteria isolated, two strains of Bacillus pacificus and Bacillus wiedmannii showed double and triple degradation efficiency for methyl red compared to the positive control (strain S12). This strategy will have good application prospects for finding new species or high-activity species of specific functional bacteria.
Keywords: bioimaging; environmental samples; functional bacteria; graphene quantum dots; single-cell in situ sorting.
Copyright © 2021 Luo, Liu, Song, Luo, Yang, Mei, Xu and Liao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
FRET-based fluorescent nanoprobe platform for sorting of active microorganisms by functional properties.Biosens Bioelectron. 2020 Jan 15;148:111832. doi: 10.1016/j.bios.2019.111832. Epub 2019 Nov 1. Biosens Bioelectron. 2020. PMID: 31706173
-
Negatively charged molybdate mediated nitrogen-doped graphene quantum dots as a fluorescence turn on probe for phosphate ion in aqueous media and living cells.Anal Chim Acta. 2019 Nov 8;1080:196-205. doi: 10.1016/j.aca.2019.07.023. Epub 2019 Jul 12. Anal Chim Acta. 2019. PMID: 31409470
-
One-step synthesis of boron-doped graphene quantum dots for fluorescent sensors and biosensor.Talanta. 2019 Jul 1;199:581-589. doi: 10.1016/j.talanta.2019.02.098. Epub 2019 Mar 1. Talanta. 2019. PMID: 30952301
-
Nitrogen-doped graphene and graphene quantum dots: A review onsynthesis and applications in energy, sensors and environment.Adv Colloid Interface Sci. 2018 Sep;259:44-64. doi: 10.1016/j.cis.2018.07.001. Epub 2018 Jul 19. Adv Colloid Interface Sci. 2018. PMID: 30032930 Review.
-
Recent Advances in the Cancer Bioimaging with Graphene Quantum Dots.Curr Med Chem. 2018;25(25):2876-2893. doi: 10.2174/0929867324666170223154145. Curr Med Chem. 2018. PMID: 28240167 Review.
Cited by
-
Primary Amine Functionalized Carbon Dots for Dead and Alive Bacterial Imaging.Nanomaterials (Basel). 2023 Jan 21;13(3):437. doi: 10.3390/nano13030437. Nanomaterials (Basel). 2023. PMID: 36770398 Free PMC article.
-
Environmental community transcriptomics: strategies and struggles.Brief Funct Genomics. 2025 Jan 15;24:elae033. doi: 10.1093/bfgp/elae033. Brief Funct Genomics. 2025. PMID: 39183066 Free PMC article. Review.
-
Bioresource-Functionalized Quantum Dots for Energy Generation and Storage: Recent Advances and Feature Perspective.Nanomaterials (Basel). 2022 Nov 5;12(21):3905. doi: 10.3390/nano12213905. Nanomaterials (Basel). 2022. PMID: 36364683 Free PMC article. Review.
References
-
- Anawar H. M., Chowdhury R. (2020). Remediation of polluted river water by biological, chemical, ecological and engineering processes. Sustainability 12:7017. 10.3390/su12177017 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

