Characterization of Human CD4 T Cells Specific for a C-Peptide/C-Peptide Hybrid Insulin Peptide
- PMID: 34113344
- PMCID: PMC8185328
- DOI: 10.3389/fimmu.2021.668680
Characterization of Human CD4 T Cells Specific for a C-Peptide/C-Peptide Hybrid Insulin Peptide
Abstract
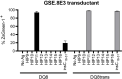
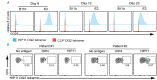
Hybrid Insulin Peptides (HIPs), which consist of insulin fragments fused to other peptides from β-cell secretory granule proteins, are CD4 T cell autoantigens in type 1 diabetes (T1D). We have studied HIPs and HIP-reactive CD4 T cells extensively in the context of the non-obese diabetic (NOD) mouse model of autoimmune diabetes and have shown that CD4 T cells specific for HIPs are major contributors to disease pathogenesis. Additionally, in the human context, HIP-reactive CD4 T cells can be found in the islets and peripheral blood of T1D patients. Here, we performed an in-depth characterization of the CD4 T cell response to a C-peptide/C-peptide HIP (HIP11) in human T1D. We identified the TCR expressed by the previously-reported HIP11-reactive CD4 T cell clone E2, which was isolated from the peripheral blood of a T1D patient, and determined that it recognizes HIP11 in the context of HLA-DQ2. We also identified a HIP11-specific TCR directly in the islets of a T1D donor and demonstrated that this TCR recognizes a different minimal epitope of HIP11 presented by HLA-DQ8. We generated and tested an HLA-DQ2 tetramer loaded with HIP11 that will enable direct ex vivo interrogation of CD4 T cell responses to HIP11 in human patients and control subjects. Using mass spectrometric analysis, we confirmed that HIP11 is present in human islets. This work represents an important step in characterizing the role of CD4 T cell responses to HIPs in human T1D.
Keywords: CD4; autoimmunity; hybrid insulin peptides; mass spectrometry; neoepitope; tetramer; type 1 diabetes.
Copyright © 2021 Wiles, Hohenstein, Landry, Dang, Powell, Guyer, James, Nakayama, Haskins, Delong and Baker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

