Human colorectal cancer-on-chip model to study the microenvironmental influence on early metastatic spread
- PMID: 34113836
- PMCID: PMC8169959
- DOI: 10.1016/j.isci.2021.102509
Human colorectal cancer-on-chip model to study the microenvironmental influence on early metastatic spread
Abstract
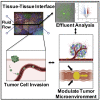
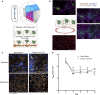
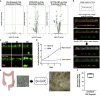
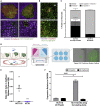
Colorectal cancer (CRC) progression is a complex process that is not well understood. We describe an in vitro organ-on-chip model that emulates in vivo tissue structure and the tumor microenvironment (TME) to better understand intravasation, an early step in metastasis. The CRC-on-chip incorporates fluid flow and peristalsis-like cyclic stretching and consists of endothelial and epithelial compartments, separated by a porous membrane. On-chip imaging and effluent analyses are used to interrogate CRC progression and the resulting cellular heterogeneity. Mass spectrometry-based metabolite profiles are indicative of a CRC disease state. Tumor cells intravasate from the epithelial channel to the endothelial channel, revealing differences in invasion between aggressive and non-aggressive tumor cells. Tuning the TME by peristalsis-like mechanical forces, the epithelial:endothelial interface, and the addition of fibroblasts influences the invasive capabilities of tumor cells. The CRC-on-chip is a tunable human-relevant model system and a valuable tool to study early invasive events in cancer.
Keywords: Bioengineering; Biomedical materials; Cancer; Classification Description; Tissue engineering.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Basson M.D. Effects of repetitive deformation on intestinal epithelial cells. Inflammopharmacology. 2007;15:109–114. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

