Managing Experimental 3D Structures in the Beyond-Rule-of-5 Chemical Space: The Case of Rifampicin
- PMID: 34114271
- PMCID: PMC8361677
- DOI: 10.1002/chem.202100961
Managing Experimental 3D Structures in the Beyond-Rule-of-5 Chemical Space: The Case of Rifampicin
Abstract
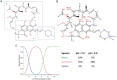
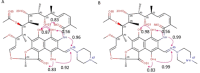
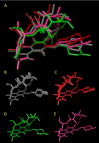
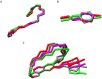
The beyond-Rule-of-5 (bRo5) chemical space is a source of new oral drugs and includes large and flexible compounds. Because of their size and conformational variability, bRo5 molecules assume different privileged conformations in the compartments of human body, i. e., they can exhibit chameleonic properties. The elucidation of the ensemble of 3D structures explored by such molecules under different conditions is therefore critical to check the role played by chameleonicity to modulate cell permeability. Here we characterized the conformational ensembles of rifampicin, a bRo5 drug, in polar and nonpolar solvents and in the solid state. We performed NMR experiments, analyzed their results with a novel algorithm and set-up a pool of ad hoc in silico strategies to investigate crystallographic structures retrieved from the CSD. Moreover, a polarity descriptor often related to permeability (SA-3D-PSA) was calculated for all the conformers and its variation with the environment analyzed. Results showed that the conformational behavior of rifampicin in solution and in the solid state is not superposable. The identification of dynamic intramolecular hydrogen bonds can be assessed by NMR spectroscopy but not by X-ray structures. Moreover, SA-3D-PSA revealed that dynamic IMHBs do not provide rifampicin with chameleonic properties. Overall, this study highlights that the peculiarity of rifampicin, which is cell permeable probably because of the presence of static IMHBs but is devoid of any chameleonic behavior, can be assessed by a proper analysis of experimental 3D structures.
Keywords: NMR spectroscopy; bRo5; chameleonicity; intramolecular hydrogen bonds; macrocycles.
© 2021 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



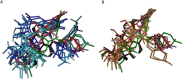
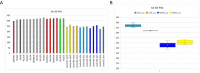
References
-
- Doak B. C., Over B., Giordanetto F., Kihlberg J., Chem. Biol. 2014, 21, 1115–42. - PubMed
-
- Poongavanam V., Doak B. C., Kihlberg J., Curr. Opin. Chem. Biol. 2018, 44, 23–29. - PubMed
-
- Ermondi G., Vallaro M., Goetz G. H., Shalaeva M., Caron G., Eur. J. Pharm. Sci. 2020, 146, 105274. - PubMed
-
- Rossi Sebastiano M., Doak B. C. B. C., Backlund M., Poongavanam V., Over B., Ermondi G., Caron G., Matsson P., Kihlberg J., J. Med. Chem. 2018, 61, 4189–4202. - PubMed

