Dual role of EZH2 in megakaryocyte differentiation
- PMID: 34115825
- PMCID: PMC8554649
- DOI: 10.1182/blood.2019004638
Dual role of EZH2 in megakaryocyte differentiation
Abstract
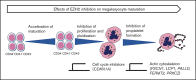
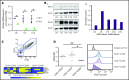
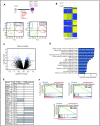
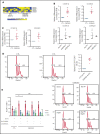
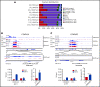
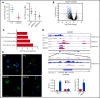
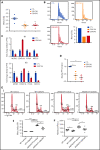
EZH2, the enzymatic component of PRC2, has been identified as a key factor in hematopoiesis. EZH2 loss-of-function mutations have been found in myeloproliferative neoplasms, particularly in myelofibrosis, but the precise function of EZH2 in megakaryopoiesis is not fully delineated. Here, we show that EZH2 inhibition by small molecules and short hairpin RNA induces megakaryocyte (MK) commitment by accelerating lineage marker acquisition without change in proliferation. Later in differentiation, EZH2 inhibition blocks proliferation and polyploidization and decreases proplatelet formation. EZH2 inhibitors similarly reduce MK polyploidization and proplatelet formation in vitro and platelet levels in vivo in a JAK2V617F background. In transcriptome profiling, the defect in proplatelet formation was associated with an aberrant actin cytoskeleton regulation pathway, whereas polyploidization was associated with an inhibition of expression of genes involved in DNA replication and repair and an upregulation of cyclin-dependent kinase inhibitors, particularly CDKN1A and CDKN2D. The knockdown of CDKN1A and to a lesser extent CDKN2D could partially rescue the percentage of polyploid MKs. Moreover, H3K27me3 and EZH2 chromatin immunoprecipitation assays revealed that CDKN1A is a direct EZH2 target and CDKN2D expression is not directly regulated by EZH2, suggesting that EZH2 controls MK polyploidization directly through CDKN1A and indirectly through CDKN2D.
© 2021 by The American Society of Hematology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

