Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation
- PMID: 34115964
- PMCID: PMC8189765
- DOI: 10.1016/j.immuni.2021.05.004
Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation
Abstract
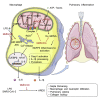
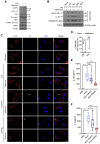
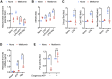
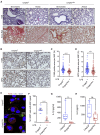
Acute respiratory distress syndrome (ARDS), an inflammatory condition with high mortality rates, is common in severe COVID-19, whose risk is reduced by metformin rather than other anti-diabetic medications. Detecting of inflammasome assembly in post-mortem COVID-19 lungs, we asked whether and how metformin inhibits inflammasome activation while exerting its anti-inflammatory effect. We show that metformin inhibited NLRP3 inflammasome activation and interleukin (IL)-1β production in cultured and alveolar macrophages along with inflammasome-independent IL-6 secretion, thus attenuating lipopolysaccharide (LPS)- and SARS-CoV-2-induced ARDS. By targeting electron transport chain complex 1 and independently of AMP-activated protein kinase (AMPK) or NF-κB, metformin blocked LPS-induced and ATP-dependent mitochondrial (mt) DNA synthesis and generation of oxidized mtDNA, an NLRP3 ligand. Myeloid-specific ablation of LPS-induced cytidine monophosphate kinase 2 (CMPK2), which is rate limiting for mtDNA synthesis, reduced ARDS severity without a direct effect on IL-6. Thus, inhibition of ATP and mtDNA synthesis is sufficient for ARDS amelioration.
Keywords: ARDS; CMPK2; COVID-19; IL-1β; IL-6; NLRP3 inflammasome; SARS-CoV-2; inflammation; metformin; mitochondrial DNA.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.K. is a founder of Elgia Pharmaceuticals and receives research support from Gossamer Bio, Jansen Pharmaceuticals and Merck. G.R.A.-M., G.L., and P.S. are employees of Jansen Pharmaceuticals. The University of California San Diego is in the process of applying for a patent covering the generation and use of novel anti-inflammatory therapy for ARDS listing H.X., E.S.-L., and M.K. as inventors. All other authors declare no competing interests.
Figures




References
-
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

