Microscopic examination of spatial transcriptome using Seq-Scope
- PMID: 34115981
- PMCID: PMC8238917
- DOI: 10.1016/j.cell.2021.05.010
Microscopic examination of spatial transcriptome using Seq-Scope
Abstract
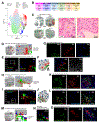
Spatial barcoding technologies have the potential to reveal histological details of transcriptomic profiles; however, they are currently limited by their low resolution. Here, we report Seq-Scope, a spatial barcoding technology with a resolution comparable to an optical microscope. Seq-Scope is based on a solid-phase amplification of randomly barcoded single-molecule oligonucleotides using an Illumina sequencing platform. The resulting clusters annotated with spatial coordinates are processed to expose RNA-capture moiety. These RNA-capturing barcoded clusters define the pixels of Seq-Scope that are ∼0.5-0.8 μm apart from each other. From tissue sections, Seq-Scope visualizes spatial transcriptome heterogeneity at multiple histological scales, including tissue zonation according to the portal-central (liver), crypt-surface (colon) and inflammation-fibrosis (injured liver) axes, cellular components including single-cell types and subtypes, and subcellular architectures of nucleus and cytoplasm. Seq-Scope is quick, straightforward, precise, and easy-to-implement and makes spatial single-cell analysis accessible to a wide group of biomedical researchers.
Keywords: RNA capture; Spatial transcriptomics; colon; high resolution; histology; liver; molecular barcoding; scRNA-seq; spatial single cell analysis; subcellular analysis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.H.L. is an inventor on pending patent applications related to Seq-Scope. H.M.K. is presently an employee of Regeneron Pharmaceuticals, in which he owns stock and stock options.
Figures
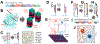





References
-
- Asp M, Bergenstrahle J, and Lundeberg J (2020). Spatially Resolved Transcriptomes-Next Generation Tools for Tissue Exploration. Bioessays 42, e1900221. - PubMed
-
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37, 38–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK114131/DK/NIDDK NIH HHS/United States
- T32 AG000114/AG/NIA NIH HHS/United States
- R01 DK102850/DK/NIDDK NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 AG024824/AG/NIA NIH HHS/United States
- K01 AG061236/AG/NIA NIH HHS/United States
- R01 DK118631/DK/NIDDK NIH HHS/United States
- P30 DK089503/DK/NIDDK NIH HHS/United States
- P30 DK034933/DK/NIDDK NIH HHS/United States
- P30 AR069620/AR/NIAMS NIH HHS/United States
- R03 HD098552/HD/NICHD NIH HHS/United States
- U01 HL137182/HL/NHLBI NIH HHS/United States
- U2C DK110768/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

