Perspectives on formaldehyde dysregulation: Mitochondrial DNA damage and repair in mammalian cells
- PMID: 34116475
- PMCID: PMC9014805
- DOI: 10.1016/j.dnarep.2021.103134
Perspectives on formaldehyde dysregulation: Mitochondrial DNA damage and repair in mammalian cells
Abstract
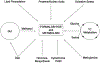
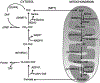
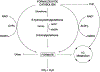
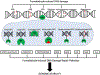
Maintaining genome stability involves coordination between different subcellular compartments providing cells with DNA repair systems that safeguard against environmental and endogenous stresses. Organisms produce the chemically reactive molecule formaldehyde as a component of one-carbon metabolism, and cells maintain systems to regulate endogenous levels of formaldehyde under physiological conditions, preventing genotoxicity, among other adverse effects. Dysregulation of formaldehyde is associated with several diseases, including cancer and neurodegenerative disorders. In the present review, we discuss the complex topic of endogenous formaldehyde metabolism and summarize advances in research on fo dysregulation, along with future research perspectives.
Keywords: DNA damage; Formaldehyde; Mitochondrial DNA; One carbon metabolism.
Copyright © 2021. Published by Elsevier B.V.
Figures

Similar articles
-
Mitochondrial dysfunction and DNA damage accompany enhanced levels of formaldehyde in cultured primary human fibroblasts.Sci Rep. 2020 Mar 27;10(1):5575. doi: 10.1038/s41598-020-61477-2. Sci Rep. 2020. PMID: 32221313 Free PMC article.
-
Review of the genotoxicity of formaldehyde.Mutat Res. 1988 Jul;196(1):37-59. doi: 10.1016/0165-1110(88)90027-9. Mutat Res. 1988. PMID: 3292899 Review. No abstract available.
-
The Role of DNA Repair in Maintaining Mitochondrial DNA Stability.Adv Exp Med Biol. 2017;1038:85-105. doi: 10.1007/978-981-10-6674-0_7. Adv Exp Med Biol. 2017. PMID: 29178071 Review.
-
Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism.Nature. 2017 Aug 31;548(7669):549-554. doi: 10.1038/nature23481. Epub 2017 Aug 16. Nature. 2017. PMID: 28813411 Free PMC article.
-
DNA damage related crosstalk between the nucleus and mitochondria.Free Radic Biol Med. 2017 Jun;107:216-227. doi: 10.1016/j.freeradbiomed.2016.11.050. Epub 2016 Nov 30. Free Radic Biol Med. 2017. PMID: 27915046 Free PMC article. Review.
Cited by
-
The Protective Effect of Mangiferin on Formaldehyde-Induced HT22 Cell Damage and Cognitive Impairment.Pharmaceutics. 2023 May 23;15(6):1568. doi: 10.3390/pharmaceutics15061568. Pharmaceutics. 2023. PMID: 37376018 Free PMC article.
-
Electrochemical and Optical Sensors for the Detection of Chemical Carcinogens Causing Leukemia.Sensors (Basel). 2023 Mar 23;23(7):3369. doi: 10.3390/s23073369. Sensors (Basel). 2023. PMID: 37050429 Free PMC article. Review.
-
Ethanol Metabolism and Melanoma.Cancers (Basel). 2023 Feb 16;15(4):1258. doi: 10.3390/cancers15041258. Cancers (Basel). 2023. PMID: 36831600 Free PMC article. Review.
-
Introducing the Role of Genotoxicity in Neurodegenerative Diseases and Neuropsychiatric Disorders.Int J Mol Sci. 2024 Jun 29;25(13):7221. doi: 10.3390/ijms25137221. Int J Mol Sci. 2024. PMID: 39000326 Free PMC article. Review.
-
Systematic Screening of Trigger Moieties for Designing Formaldehyde Fluorescent Probes and Application in Live Cell Imaging.Biosensors (Basel). 2022 Oct 10;12(10):855. doi: 10.3390/bios12100855. Biosensors (Basel). 2022. PMID: 36290992 Free PMC article.
References
-
- Li E, Bestor TH, Jaenisch R, Targeted mutation of the DNA methyltransferase gene results in embryonic lethality, Cell, 69 (1992) 915–926. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E, DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development, Cell, 99 (1999) 247–257. - PubMed
-
- MacAllister SL, Choi J, Dedina L, O'Brien PJ, Metabolic mechanisms of methanol/formaldehyde in isolated rat hepatocytes: carbonyl-metabolizing enzymes versus oxidative stress, Chem. Biol. Interact, 191 (2011) 308–314. - PubMed
-
- Dorokhov YL, Shindyapina AV, Sheshukova EV, Komarova TV, Metabolic methanol: molecular pathways and physiological roles, Physiol. Rev, 95 (2015) 603–644. - PubMed
-
- Diliberto EJ, Axelrod J, Regional and subcellular distribution of protein carboxymethylase in brain and other tissues, J. Neurochem, 26 (1976) 1159–1165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

