ADP-ribosylation of DNA and RNA
- PMID: 34116477
- PMCID: PMC8385414
- DOI: 10.1016/j.dnarep.2021.103144
ADP-ribosylation of DNA and RNA
Abstract
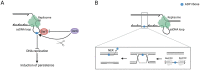
ADP-ribosylation is a chemical modification of macromolecules found across all domains of life and known to regulate a variety of cellular processes. Notably, it has a well-established role in the DNA damage response. While it was historically known as a post-translational modification of proteins, recent studies have shown that nucleic acids can also serve as substrates of reversible ADP-ribosylation. More precisely, ADP-ribosylation of DNA bases, phosphorylated DNA ends and phosphorylated RNA ends have been reported. We will discuss these three types of modification in details. In a variety of bacterial species, including Mycobacterium tuberculosis, ADP-ribosylation of thymidine has emerged as the mode of action of a toxin-antitoxin system named DarTG, with the resultant products perceived as DNA damage by the cell. On the other hand, mammalian DNA damage sensors PARP1, PARP2 and PARP3 were shown to ADP-ribosylate phosphorylated ends of double-stranded DNA in vitro. Additionally, TRPT1 and several PARP enzymes, including PARP10, can add ADP-ribose to the 5'-phosphorylated end of single-stranded RNA in vitro, representing a novel RNA capping mechanism. Together, these discoveries have led to the emergence of a new and exciting research area, namely DNA and RNA ADP-ribosylation, that is likely to have far-reaching implications for the fields of DNA repair, replication and epigenetics.
Keywords: ADP-ribosylation; DNA damage response; DNA modification; PARP; RNA modification.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

