Clinical and Functional Characterization of Atypical KRAS/ NRAS Mutations in Metastatic Colorectal Cancer
- PMID: 34117033
- PMCID: PMC8364867
- DOI: 10.1158/1078-0432.CCR-21-0180
Clinical and Functional Characterization of Atypical KRAS/ NRAS Mutations in Metastatic Colorectal Cancer
Abstract
Purpose: Mutations in KRAS/NRAS (RAS) predict lack of anti-EGFR efficacy in metastatic colorectal cancer (mCRC). However, it is unclear if all RAS mutations have similar impact, and atypical mutations beyond those in standard guidelines exist.
Experimental design: We reviewed 7 tissue and 1 cell-free DNA cohorts of 9,485 patients to characterize atypical RAS variants. Using an in vitro cell-based assay (functional annotation for cancer treatment), Ba/F3 transformation, and in vivo xenograft models of transduced isogenic clones, we assessed signaling changes across mutations.
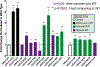
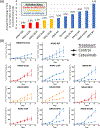
Results: KRAS exon 2, extended RAS, and atypical RAS mutations were noted in 37.8%, 9.5%, and 1.2% of patients, respectively. Among atypical variants, KRAS L19F, Q22K, and D33E occurred at prevalence ≥0.1%, whereas no NRAS codon 117/146 and only one NRAS codon 59 mutation was noted. Atypical RAS mutations had worse overall survival than RAS/BRAF wild-type mCRC (HR, 2.90; 95% confidence interval, 1.24-6.80; P = 0.014). We functionally characterized 114 variants with the FACT assay. All KRAS exon 2 and extended RAS mutations appeared activating. Of 57 atypical RAS variants characterized, 18 (31.6%) had signaling below wild-type, 23 (40.4%) had signaling between wild-type and activating control, and 16 (28.1%) were hyperactive beyond the activating control. Ba/F3 transformation (17/18 variants) and xenograft model (7/8 variants) validation was highly concordant with FACT results, and activating atypical variants were those that occurred at highest prevalence in clinical cohorts.
Conclusions: We provide best available evidence to guide treatment when atypical RAS variants are identified. KRAS L19F, Q22K, D33E, and T50I are more prevalent than many guideline-included RAS variants and functionally relevant.
©2021 American Association for Cancer Research.
Figures



References
-
- Peeters M, Kafatos G, Taylor A, Gastanaga VM, Oliner KS, Hechmati G, et al.Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer. 2015;51:1704–13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26049686 - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al.Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. - PubMed
-
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al.K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. - PubMed
-
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al.Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:1346–55. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24718886 - PubMed
-
- Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al.Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24024839 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

