Distinct clinical and immunological profiles of patients with evidence of SARS-CoV-2 infection in sub-Saharan Africa
- PMID: 34117221
- PMCID: PMC8196064
- DOI: 10.1038/s41467-021-23267-w
Distinct clinical and immunological profiles of patients with evidence of SARS-CoV-2 infection in sub-Saharan Africa
Abstract
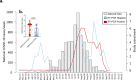
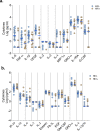
Although the COVID-19 pandemic has left no country untouched there has been limited research to understand clinical and immunological responses in African populations. Here we characterise patients hospitalised with suspected (PCR-negative/IgG-positive) or confirmed (PCR-positive) COVID-19, and healthy community controls (PCR-negative/IgG-negative). PCR-positive COVID-19 participants were more likely to receive dexamethasone and a beta-lactam antibiotic, and survive to hospital discharge than PCR-negative/IgG-positive and PCR-negative/IgG-negative participants. PCR-negative/IgG-positive participants exhibited a nasal and systemic cytokine signature analogous to PCR-positive COVID-19 participants, predominated by chemokines and neutrophils and distinct from PCR-negative/IgG-negative participants. PCR-negative/IgG-positive participants had increased propensity for Staphylococcus aureus and Streptococcus pneumoniae colonisation. PCR-negative/IgG-positive individuals with high COVID-19 clinical suspicion had inflammatory profiles analogous to PCR-confirmed disease and potentially represent a target population for COVID-19 treatment strategies.
Conflict of interest statement
The authors declare no competing interests
Figures



Update of
-
In depth analysis of patients with severe SARS-CoV-2 in sub-Saharan Africa demonstrates distinct clinical and immunological profiles.medRxiv [Preprint]. 2021 Feb 20:2021.02.15.21251753. doi: 10.1101/2021.02.15.21251753. medRxiv. 2021. Update in: Nat Commun. 2021 Jun 11;12(1):3554. doi: 10.1038/s41467-021-23267-w. PMID: 33619502 Free PMC article. Updated. Preprint.
References
-
- Government S. A. Update on Covid-19. https://sacoronavirus.co.za (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

