The functional organization of excitatory synaptic input to place cells
- PMID: 34117238
- PMCID: PMC8196201
- DOI: 10.1038/s41467-021-23829-y
The functional organization of excitatory synaptic input to place cells
Abstract
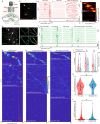
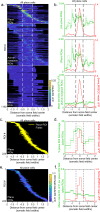
Hippocampal place cells contribute to mammalian spatial navigation and memory formation. Numerous models have been proposed to explain the location-specific firing of this cognitive representation, but the pattern of excitatory synaptic input leading to place firing is unknown, leaving no synaptic-scale explanation of place coding. Here we used resonant scanning two-photon microscopy to establish the pattern of synaptic glutamate input received by CA1 place cells in behaving mice. During traversals of the somatic place field, we found increased excitatory dendritic input, mainly arising from inputs with spatial tuning overlapping the somatic field, and functional clustering of this input along the dendrites over ~10 µm. These results implicate increases in total excitatory input and co-activation of anatomically clustered synaptic input in place firing. Since they largely inherit their fields from upstream synaptic partners with similar fields, many CA1 place cells appear to be part of multi-brain-region cell assemblies forming representations of specific locations.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Zhao, X., Wang, Y., Spruston, N. & Magee, J. C. Membrane potential dynamics underlying context-dependent sensory responses in the hippocampus. Nat. Neurosci.23, 881–891 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

