Cas9 targeted enrichment of mobile elements using nanopore sequencing
- PMID: 34117247
- PMCID: PMC8196195
- DOI: 10.1038/s41467-021-23918-y
Cas9 targeted enrichment of mobile elements using nanopore sequencing
Abstract
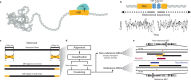
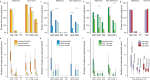
Mobile element insertions (MEIs) are repetitive genomic sequences that contribute to genetic variation and can lead to genetic disorders. Targeted and whole-genome approaches using short-read sequencing have been developed to identify reference and non-reference MEIs; however, the read length hampers detection of these elements in complex genomic regions. Here, we pair Cas9-targeted nanopore sequencing with computational methodologies to capture active MEIs in human genomes. We demonstrate parallel enrichment for distinct classes of MEIs, averaging 44% of reads on-targeted signals and exhibiting a 13.4-54x enrichment over whole-genome approaches. We show an individual flow cell can recover most MEIs (97% L1Hs, 93% AluYb, 51% AluYa, 99% SVA_F, and 65% SVA_E). We identify seventeen non-reference MEIs in GM12878 overlooked by modern, long-read analysis pipelines, primarily in repetitive genomic regions. This work introduces the utility of nanopore sequencing for MEI enrichment and lays the foundation for rapid discovery of elusive, repetitive genetic elements.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

