Quantitative model of eukaryotic Cdk control through the Forkhead CONTROLLER
- PMID: 34117265
- PMCID: PMC8196193
- DOI: 10.1038/s41540-021-00187-5
Quantitative model of eukaryotic Cdk control through the Forkhead CONTROLLER
Abstract
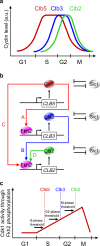
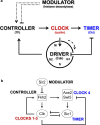
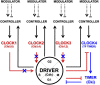
In budding yeast, synchronization of waves of mitotic cyclins that activate the Cdk1 kinase occur through Forkhead transcription factors. These molecules act as controllers of their sequential order and may account for the separation in time of incompatible processes. Here, a Forkhead-mediated design principle underlying the quantitative model of Cdk control is proposed for budding yeast. This design rationalizes timing of cell division, through progressive and coordinated cyclin/Cdk-mediated phosphorylation of Forkhead, and autonomous cyclin/Cdk oscillations. A "clock unit" incorporating this design that regulates timing of cell division is proposed for both yeast and mammals, and has a DRIVER operating the incompatible processes that is instructed by multiple CLOCKS. TIMERS determine whether the clocks are active, whereas CONTROLLERS determine how quickly the clocks shall function depending on external MODULATORS. This "clock unit" may coordinate temporal waves of cyclin/Cdk concentration/activity in the eukaryotic cell cycle making the driver operate the incompatible processes, at separate times.
Conflict of interest statement
The author declares no competing interests.
Figures

Similar articles
-
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast.NPJ Syst Biol Appl. 2021 Dec 13;7(1):48. doi: 10.1038/s41540-021-00201-w. NPJ Syst Biol Appl. 2021. PMID: 34903735 Free PMC article. Review.
-
Evidence of substrate control of Cdk phosphorylation during the budding yeast cell cycle.Cell Rep. 2025 Apr 22;44(4):115534. doi: 10.1016/j.celrep.2025.115534. Epub 2025 Apr 11. Cell Rep. 2025. PMID: 40220295
-
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].Cancer Radiother. 2001 Apr;5(2):109-29. doi: 10.1016/s1278-3218(01)00087-7. Cancer Radiother. 2001. PMID: 11355576 Review. French.
-
Clb3-centered regulations are recurrent across distinct parameter regions in minimal autonomous cell cycle oscillator designs.NPJ Syst Biol Appl. 2020 Apr 3;6(1):8. doi: 10.1038/s41540-020-0125-0. NPJ Syst Biol Appl. 2020. PMID: 32245958 Free PMC article.
-
The cyclin family of budding yeast: abundant use of a good idea.Trends Genet. 1998 Feb;14(2):66-72. doi: 10.1016/s0168-9525(97)01322-x. Trends Genet. 1998. PMID: 9520600 Review.
Cited by
-
A yeast cell cycle model integrating stress, signaling, and physiology.FEMS Yeast Res. 2022 Jun 30;22(1):foac026. doi: 10.1093/femsyr/foac026. FEMS Yeast Res. 2022. PMID: 35617157 Free PMC article.
-
Metabolic imbalance driving immune cell phenotype switching in autoimmune disorders: Tipping the balance of T- and B-cell interactions.Clin Transl Med. 2024 Mar;14(3):e1626. doi: 10.1002/ctm2.1626. Clin Transl Med. 2024. PMID: 38500390 Free PMC article.
-
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast.NPJ Syst Biol Appl. 2021 Dec 13;7(1):48. doi: 10.1038/s41540-021-00201-w. NPJ Syst Biol Appl. 2021. PMID: 34903735 Free PMC article. Review.
-
Unveiling Forkhead-mediated regulation of yeast cell cycle and metabolic networks.Comput Struct Biotechnol J. 2022 Apr 7;20:1743-1751. doi: 10.1016/j.csbj.2022.03.033. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35495119 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

