Assessment of a complete and classified platelet proteome from genome-wide transcripts of human platelets and megakaryocytes covering platelet functions
- PMID: 34117303
- PMCID: PMC8196183
- DOI: 10.1038/s41598-021-91661-x
Assessment of a complete and classified platelet proteome from genome-wide transcripts of human platelets and megakaryocytes covering platelet functions
Abstract
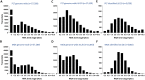
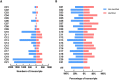
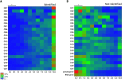
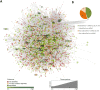
Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome-wide transcriptomes (57.8 k mRNAs). For 14.8 k protein-coding transcripts, we assigned the proteins to 21 UniProt-based classes, based on their preferential intracellular localization and presumed function. This classified transcriptome-proteome profile of platelets revealed: (i) Absence of 37.2 k genome-wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein-coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43-0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identified proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma-derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identified proteome of nuclear-related, membrane and signaling proteins, as well proteins with low-level transcripts. We then constructed a prediction model, based on protein function, transcript level and (peri)nuclear localization, and calculated the achievable proteome at ~ 10 k proteins. Model validation identified 1.0 k additional proteins in the predicted classes. Network and database analysis revealed the presence of 2.4 k proteins with a possible role in thrombosis and hemostasis, and 138 proteins linked to platelet-related disorders. This genome-wide platelet transcriptome and (non)identified proteome database thus provides a scaffold for discovering the roles of unknown platelet proteins in health and disease.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Werner G, Morgenstern E. Three-dimensional reconstruction of human blood platelets using serial sections. Eur. J. Cell. Biol. 1980;20:276–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

