Non-coding RNAs in chromatin folding and nuclear organization
- PMID: 34117518
- PMCID: PMC11072467
- DOI: 10.1007/s00018-021-03876-w
Non-coding RNAs in chromatin folding and nuclear organization
Abstract
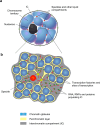
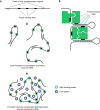
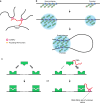
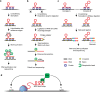
One of the most intriguing questions facing modern biology concerns how the genome directs the construction of cells, tissues, and whole organisms. It is tempting to suggest that the part of the genome that does not encode proteins contains architectural plans. We are still far from understanding how these plans work at the level of building tissues and the body as a whole. However, the results of recent studies demonstrate that at the cellular level, special non-coding RNAs serve as scaffolds for the construction of various intracellular structures. The term "architectural RNAs" was proposed to designate this subset of non-coding RNAs. In this review, we discuss the role of architectural RNAs in the construction of the cell nucleus and maintenance of the three-dimensional organization of the genome.
Keywords: 3D genome; Liquid condensate; Non-coding RNA; Nucleus; Transcription.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

